Hydrogen bonding plays a key role in structure and function of proteins including features such as protein folding, local architecture, protein-ligand recognition, enzymatic activity, protein hydration and molecular dynamics. These classical hydrogen bonds are well established and have been studied in detail (Baker and Hubbard, 1984; Jeffrey and Saenger, 1991). However, a set of somewhat weaker interactions has also been recognized to play an important role in protein structure and stability (Desiraju and Steiner, 1999). This set includes N-H...π, O-H...π, C-H...O and C-H...π. The overall stabilization energy of these interactions is much smaller (not more than a few Kcal/mol), but is still signficant.
C-H...X interaction
In recent years it has been established that a C-H group can be a hydrogen bond donor. Although considered weak in nature, the bond C-H...X (X: O, π) termed as C-H interaction is known to be distrubuted widely among protein structures. These interactions have shown to be of greater importance in proteins, and polypeptides, protein-protein, protein-ligand and drug-binding interactions.
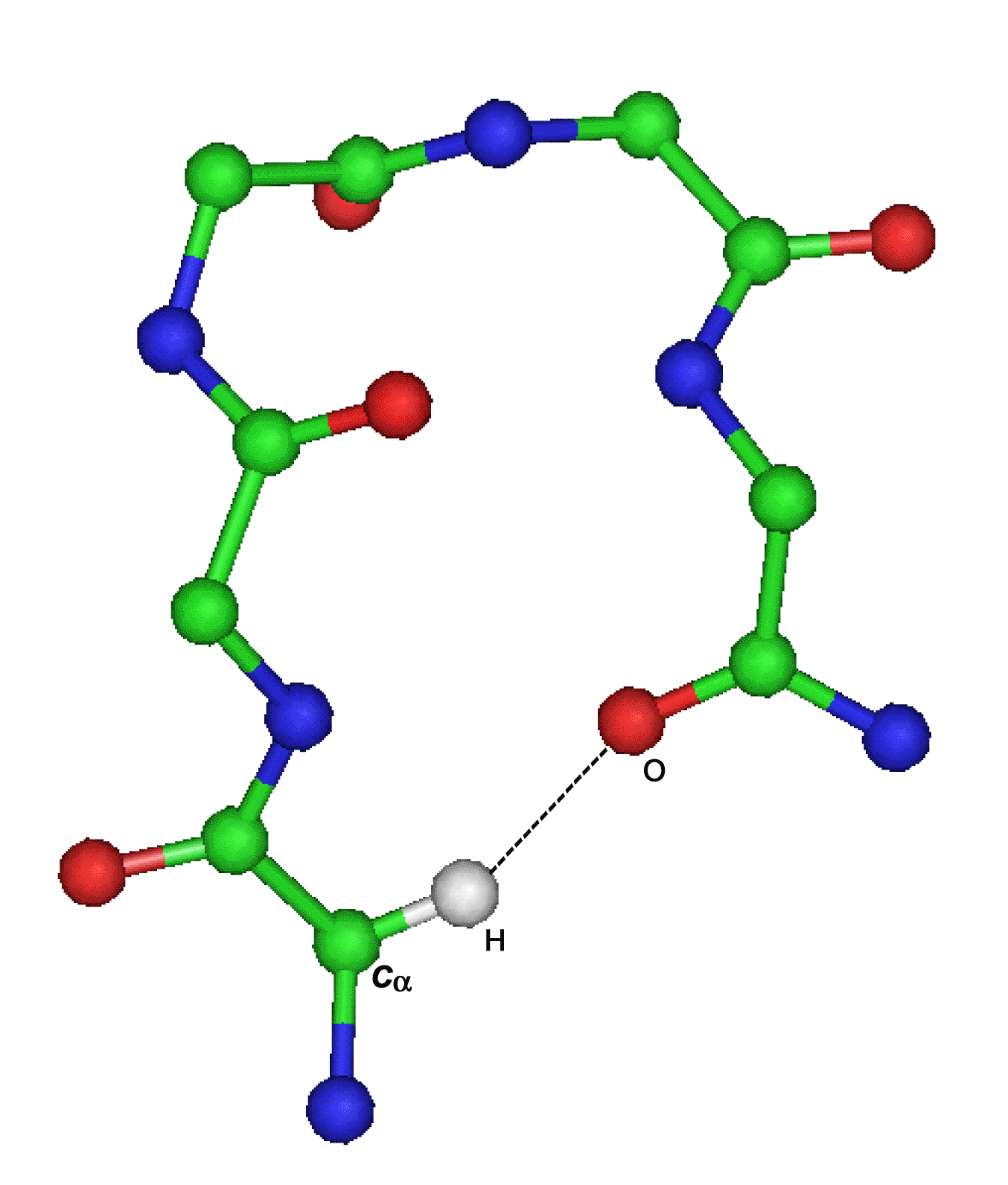

C-H...O interaction C-H...π interaction
C-H...O interaction
C-H...O interactions form 20-25% of the total number of hydrogen bonds constituting the second most important group (Weiss, 2001). Important early work indicated a role for C-H...O interactions in protein structures. Steiner and Saenger (1993) reported the presence of C-H...O bonds completing the coordination of the buried water molecules in papain. Derewenda et al. 1994 presented evidence of functional significance in serine hydrolases of a hydrogen bond involving the C-H group of the active site His and a neighboring main-chain carbonyl oxygen. The past studies have revealed that Cα-H...O hydrogen bonds occur almost ubiquitously in beta-sheets (Fabiola et al. 1997), but they also appear to occur frequently in alpha-helical proteins (Chakrabarti and Chakrabarti, 1998).
Identification of Cα-H...O interactions
The C-H...O interactions in the dataset have been identified using the program HBexplore (Lindauer et al., 1996) that requires structural information in the Protein Data Bank (PDB) format (Bernstein et al., 1977). The set I criteria with geometrical parameters (distance H...O < 2.5Α, Cα...O < 3.9Α and Cα-H...O > 90o) have been used. The program identifies all potential hydrogen bonds according to geometrical criteria and generates an output. The output lists the name, position number of donor and acceptor residues along with geometrical parameters and subgroup of the identified H-bond set e.g. backbone-backbone bond.
C-H...π interaction
Hydrogen bonds, C-H...π with π-acceptor constitute yet another considerable fraction. Steiner and Koellner, 2001 described hydrogen bonds in proteins involving aromatic acceptors, and Brandl et al. exhaustively surveyed the occurrence of interactions involving all possible C-H groups (Cα, Caliphatic-H and Caromatic-H) as donors and all possible side chain π systems as acceptors. The cases in which C-H...π interactions have been described in proteins include the formation of complexes of proteins with ligands or cofactors such as the heme group (Nishio et al. 1998) and design of serine proteases inhibitors (Shimohigashi et al. 1996, 1999). Previous studies have also shown that C-H...π interactions are even responsible for the stabilization of structural elements such as alpha or 310 helices or non-proline cis peptide bonds (Brandl et al.,2001) .
Identification of Cα-H...π interactions
The C-H...π interactions in the dataset have been identified using the web server NCI server that is based purely on geometric criteria (Babu, 2003) . The default parameters (Cα...O=C <= 3.8Α Hα...O=C <=3.3Α, Cα-H...O >=120o and Hα...O=C >= 90o) have been used. The output consists of name, position of interacting residues and the observed values for each of the parameters.
Some important references
1. Baker, E. N. & Hubbard, R.E. (1984). Hydrogen bonding in globular proteins. Prog. Biophys. Mol. Biol. 44, 97-179.[Abstract]
2. Jeffrey, G. A. & Saenger, W. (1991). Hydrogen bonding in Biological Structures, Springer-Verlag, NY.
3. Desiraju, G. & Steiner, T. (1999). The Weak Hydrogen bond in Structural Chemistry and Biology, Oxford University Press, Oxford.
4. Derewenda, Z. S., Lee, L. & Derewenda, U. (1995). The occurrence of C-H...O hydrogen bonds in proteins. J. Mol. Biol. 252, 248-262.[Abstract]
5. Brandl, M., Weiss, M. S., Jabs, A., Sühnel, J. & Hilgenfeld, R. (2001). C-H...π interactions in proteins. J. Mol. Biol. 307, 357-377.[Abstract]
6. Steiner, T. & Koellner, G. (2001). Hydrogen bonds with π-acceptors in proteins: frequencies and role in stabilizing local 3D structures. J. Mol. Biol. 305, 535-557.[Abstract]
7. Weiss, M.S. (2001). More hydrogen bonds for the (structural) biologist. Trends Biochem. Sci. 26, 521-523.[Abstract]
8. Steiner, T. & Saenger, W. (1993). The ordered water cluster in vitamin B12 coenzyme at 15K is stabilized by C-H...O hydrogen bonds. Acta Crystallog. Sect. D. 49, 592-593.
9. Desiraju, G. R. (1991). The C-H...O hydrogen bond in crystals: What is it? Acc. Chem. Res. 24, 290-296.
10. Derewenda, Z. S., Derewenda, U. & Kobos, P. (1994). (His)C-H...O=C < hydrogen bond in the active sites of serine hydrolases. J. Mol. Biol. 241, 83-93.[Abstract]
11. Fabiola, G. F., Krishnaswamy, S., Nagarajan, V., & Pattabhi, V. (1997). C-H...O hydrogen bonds in beta sheets. Acta. Cryst. D53, 316-320.
12. Chakrabarti, P & Chakrabarti S. (1998). C-H...O hydrogen bond involving proline residues in alpha-helices. J. Mol. Biol. 284, 867-873.[Abstract]
13. Babu, M. M., Singh, K. S. & Balaram, P. (2002). A C-H...O hydrogen bond stabilized polypeptide chain reversal motif at the C terminus of helices in proteins. J. Mol. Biol. 322, 871-880.[Abstract]
14. Nishio, M., Hirota, M. & Umezawa, Y. (1998). The CH/π Interaction. Evidence, Nature and Consequences. Wiley-VCH, New York.
15. Shimohigashi, Y., Maeda, I., Nose, T., Ikesue, K., Sakamoto, H., Ogawa, T., Ide, Y. & Kawahara, M., (1996). Chymotrypsin inhibitory conformation induced by amino acid side-chain side-chain intramolecular CH/π interaction. J. Chem. Soc., Perkin Trans. 1, 2479-2485.
16. Shimohigashi, Y., Nose, T., Yamauchi, Y. & Maeda, I. (1999). Design of serine protease inhibitors with conformation restricted by amino acid side-chain side-chain CH/π interaction. Biopolymers 51, 9-17.[Abstract]
17. Jones, D.T. 1999. Protein secondary structure prediction based on position specific scoring matrices. J. Mol. Biol. 292, 195-202.[Abstract]
18. Lindauer, K., Bendic, C. & Sühnel, J. (1996). HBexplore - a new tool for identifying hydrogen bonding patterns in biological macromolecules. Comput. Appl. Biosci. 12, 281-289.[Abstract]
19. Bernstein, F.C., Koetzle, T.F., Williams, G., Mayer, E.F., Bryce, M.D., Rodgers, J.R., Kennard, O., Simanouchi, T. & Tasumi, M. (1977). The Protein Data Bank: a computer based archival file for macromolecular structures. J. Mol. Biol. 112, 535-542.[Abstract]
20. Babu, M. M. (2003). NCI: a server to identify non-canonical interactions in protein structures. Nucleic Acids Res. 31, 3345-3348.[Abstract]
21. Zell, A. & Mamier, G. (1997). Stuttgart neural network simulator, version 4.2. University of Stuttgart, Stuttgart, Germany.
22. Rumelhart, D.E., Hinton, G.E. & Williams, R.J. (1986). Learning representations by back-propagation errors. Nature 323, 533-536.
