| |
|
 |
|
|
TheHLA-DR4Pred is an SVM and ANN based HLA-DRB1*0401(MHC class II alleles) binding peptides prediction method. The accuracy of the SVM and ANN based methods is ~86% and ~78% respectively.The performence of the methods was tested through 5 set cross-validation. The training of SVM and ANN was done by using the freely availaible SVM_LIGHT and SNNS packages respectively. The data for training of neural network and SVM model has been extracted from MHCBN database.This method will be useful in cellular immunology, Vaccine design, immunodiagnostics, immunotherapeuatics and molecular understanding of autoimmune susceptibility. |
|
|
|
|
|
|
|
|
|
 |
|
|
|
|
|
 General Introduction:- HLA-DR Proteins have evolved to bind and transport a wide variety of peptides obtained from the antigens to cell surface where this complex is recognized by helper T cells. In the past, a few HLA-DR proteins were found to be associated with autoimmune disease e.g. HLA-DRB1*0401 with rheumatoid arthritis . It is important for treatment of autoimmune diseases and cancer therapy to determine which peptides binds to MHC class II molecules (HLA-DR) that will help in treatment of these diseases. The experimental methods for identification of these peptides are both time consuming and cost intensive. General Introduction:- HLA-DR Proteins have evolved to bind and transport a wide variety of peptides obtained from the antigens to cell surface where this complex is recognized by helper T cells. In the past, a few HLA-DR proteins were found to be associated with autoimmune disease e.g. HLA-DRB1*0401 with rheumatoid arthritis . It is important for treatment of autoimmune diseases and cancer therapy to determine which peptides binds to MHC class II molecules (HLA-DR) that will help in treatment of these diseases. The experimental methods for identification of these peptides are both time consuming and cost intensive.
The computer-based prediction methods thus, provide a cost effective alternative . However, it is difficult to predict the peptides binding with HLA-DR molecules as compared to MHC class I molecules due to - Variable length of binding peptides.
- Undetermined core for each binding peptides.
- Range of amino acids occupying anchor position for each MHC allele.
Here, we have developed a computation tool for the prediction of HLA-DRB1*0401 binding peptides.The methods has been implemted online as HLA-DR4Pred. The prediction is based on the Support Vector Machine (SVM) and Artificial neural Network (ANN). The user can vary the threshold of prediction approach to vary the stringency of prediction. The iterative algorithm of the prediction methods is shown below.

Stepwise Algorithm and Method description:-
Extraction and Preprocessing of data:-
The MHCBN database was used as the source of data. The HLA-DRB1*0401 binding peptides of length 9 or more than 9 amino acids were obtained from this database. All the peptides with unnatural amino acids were filtered from the final dataset. The binding core of 9 aa was obtained from the binders without considering MHC binding motifs by using Matrix Optimization Techniques (MOT) package . ALL the duplicate entries were cropped from the dataset. The final dataset is consisted of 567 unique MHC binders of varying binding affinity. The HLA-DRB1*0401 non-binders of 9 or more than 9 amino acids were also obtained from MHCBN database. The non-binders were chopped into overlapping peptides of 9 amino acids. 567 unique non-binders were randomly chosen from this dataset.The final dataset is having 567 HLA-DRB1*0401 binders and 567 HLA-DRB1*0401 non-binders.
Optimization of SVM based Method:-
we have implemented a support vector machine based package SVM_LIGHT to classify the data of MHC class II binders and non-binders. The SVM is provided with an sparse input of 20 Vectors. The HLA-DRB1*0401 binders were represented by "+1" and non-binders were represnted by "-1". The suitable type of kernel for classifying the data was chosen by conducting experiments with every type of kernel i.e. RBF, Polynomial, Sigmoid and LINEAR.In case of SVM the choosing a kernel is similiar to the problem of choosing an architecture.The performance of the kernel was measured by considering the accuracy at a cutoff value where sensitivity and specificity are nearly equal. The RBF kernel was found best in classifying the data of DRB1*0401 binders and non-binders.During optimization process the other feature of the kernel g trade off value c were also fine tuned.The value regulatory parameters c and g of RBF kernel were optimized to 5.5 and 0.1 respectively.
ANN based Method:-
For neural network implementation and to generate neural network architecture and learning process, publicly available free simulation package SNNS4.2 was used.The input for artificial neural network is a single sequence in the binary representation. In this representation, each amino acid at each window position is encoded by a group of 20 input units- 20 units code for each of the possible natural amino acids at that position . In each group of 20 input units, the input corresponding to the amino acid type at that window position is set to 1 and rest all other inputs to 0. for example the alanine and glycine is represented as follows
Alanine: 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
Glycine: 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
In this manner a single peptide of 9 amino acids is represented by 180 input units to artificial
neural networks.
The training was carried out by using the standard feed-forward back propagation network with single hidden layer. The ultimate value of learning rate, learning cycles and hidden nodes were determined by monitoring the error on the training set.The performence of these machine learning based methods must be evalvuated on unseen data.The five-fold cross validation was used to verify the performance of both SVM and ANN. The dataset was divided into the five sets, each consisting of equal number of binders and non-binders. The optimized SVM model or ANN network was generated on the training set consisting of four sets and performance of method was tested on testing set. The performance of the methods were evaluated through threshold dependent parameters like sensitivity, specificity, PPV, NPV and accuracy.The value of diffent parameters optimised to
The ultimate number of cycles:300
The linear learning rate:.01
neural network function: Standard back propagation.
Number of Hidden layers:1.
Number of nodes per hidden layer=1.
Performence Measures:-
Prediction Results:-
Both the SVM and ANN based methods developed in present study performed better as compared to all other algorithms for Prediction of HLA-DRB1*0401 binding peptides. The performence of other methods is measured by implemting then on our dataset. The summary of different prediction methods on our dataset is shown in table below.
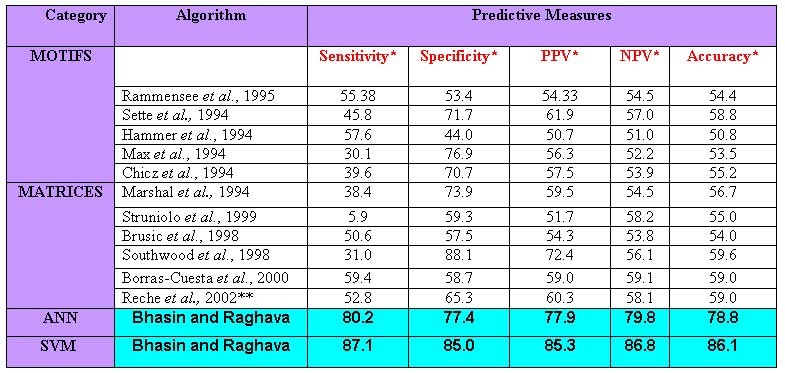
Inferences from results:- The following inferences can be obtained from the results
- The machine learning techniques based methods can classify the data more accurately as compared to motifs and matrices based methods.
- The SVM is more superior in classifying the non-linear data of MHC interacting peptides as compared to ANN.
- This observation for single MHC allele can be extended to other MHC alleles.
|
|
|