Application
QSPpred can be used for the identification, designing and analyzing new QS peptides that can be used as novel therapeutic targets.
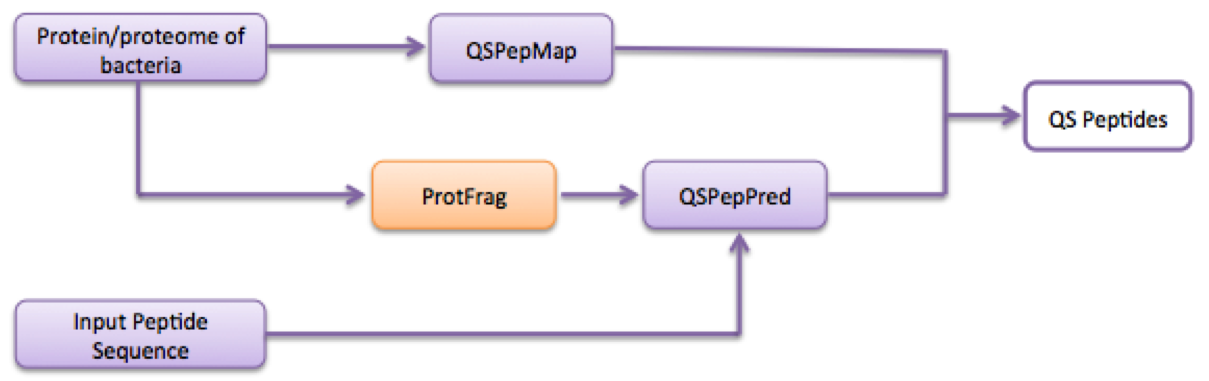
➢ If the user wants to identify new QS peptides from the proteome/protein of any clinically important bacteria (eg. Streptococcus aureus). It can be done by use of QSPpred by exploring tools and selecting different window lengths and thresholds. If the user gives peptide sequence as input and wants to check its status as QS or non-QS peptide. It can be done by the use of QSPpred webserver as given shown in flowchart:
➢ If the user want to design QS peptides by developing all possible mutants of a given peptide and to predict QS ability of designed peptides. It can be done by the use of various tools in QSPpred webserver. Flow chart of various tools to be explored in QSPpred are:
➢ If the user wants to check various possibilities from input peptide sequence i.e. which type of residues preferred at which position that gives more probability of peptide to behave as QS peptide. It can be done by the use of various tools in QSPpred webserver. Flow chart of various tools to be explored in QSPpred are:
(A) IDENTIFICATION
(B) DESIGNING

(C) ANALYZING
