Search Module |
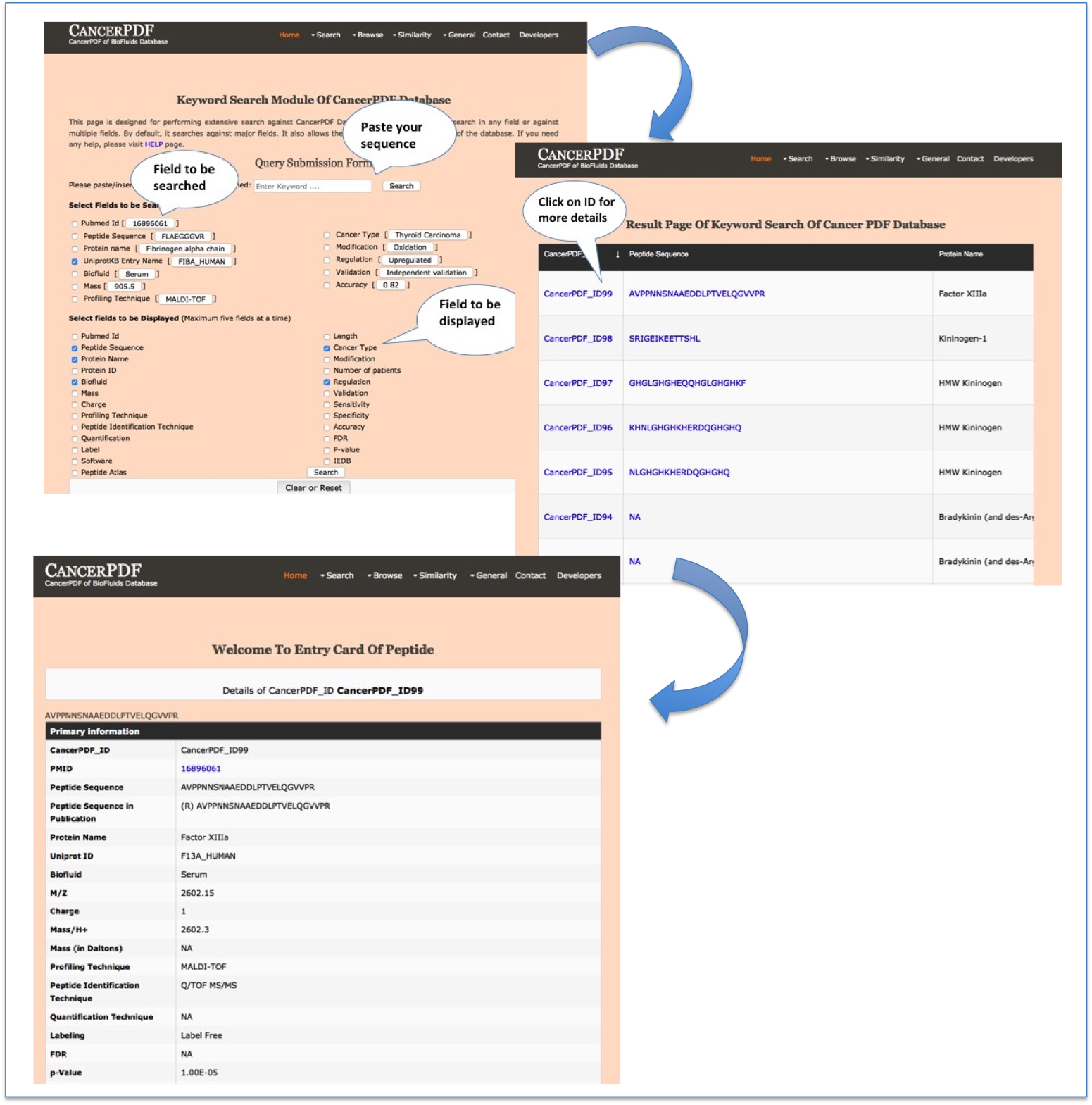
Keyword Search
This module displays the simple search page of CancerPDF. Clicking on the required field will allow the user to browse the specific information in the database. The "select fields to be searched" section provides options for various keywords through which search can be made in CancerPDF. User can select fields to be displayed from the section "fields to be displayed". User can click on each cancerPDF ID to get all the details about a entry. User can also click on each peptide to know information about that peptide.
|
|
|
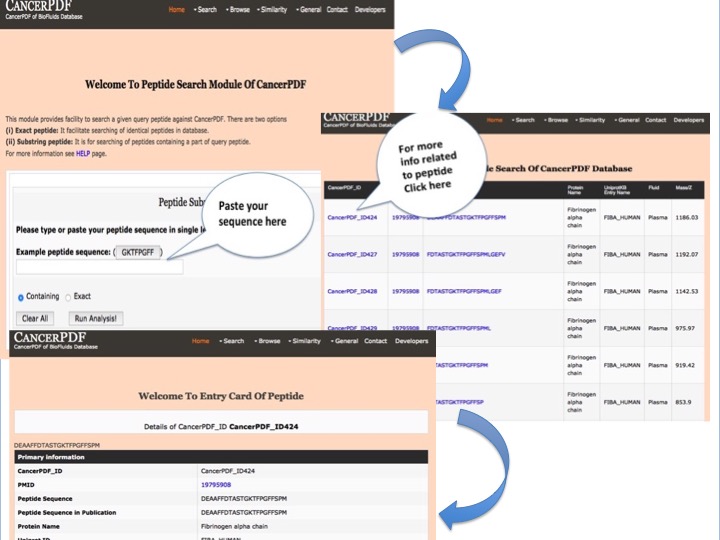
Peptide Search
This module provides facility to search a given query peptide against CancerPDF. It has two options containing and exact. Containing will fetch all the peptides that contain query peptide. Exact match will return only peptide entries that are exact to query peptide.
|
|
|
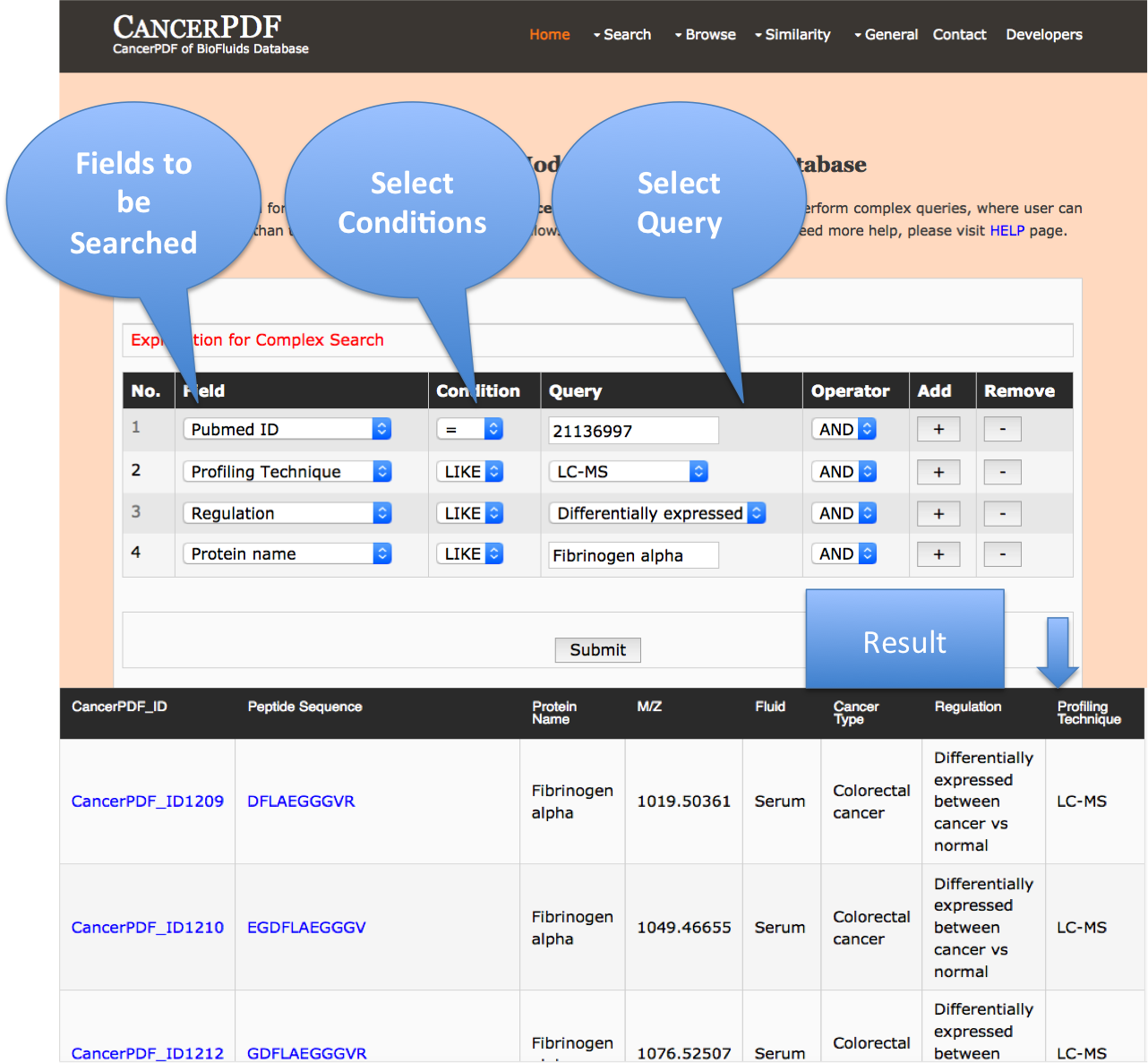
Advance Search
This page provides the advanced search facility in CancerPDF. User can browse the page by selecting two or more than two fileds.
|
| Complex Search is quite helpful when user has predefined query in mind. For instance, User come up with the following questions in mind and wants to SEARCH |
| 1. Precursor protein by name e.g. Fibrinogen Alpha. |
| 2. Among the above selected peptides, select those having LC-MS profiling technique. |
| 3. Among the above selection criteria, show peptides with category Differentially Expressed in Colorectal cancer only. |
| 4. Among the above selected peptides, select only those having atleast GEGDFLAEGGGVR residues in the sequences. |
| By clicking Submit; user can get desired output. |
| In a similar way, query can be extended using "+" button and reduced using "-" button. |
|
|
Browse page |
|
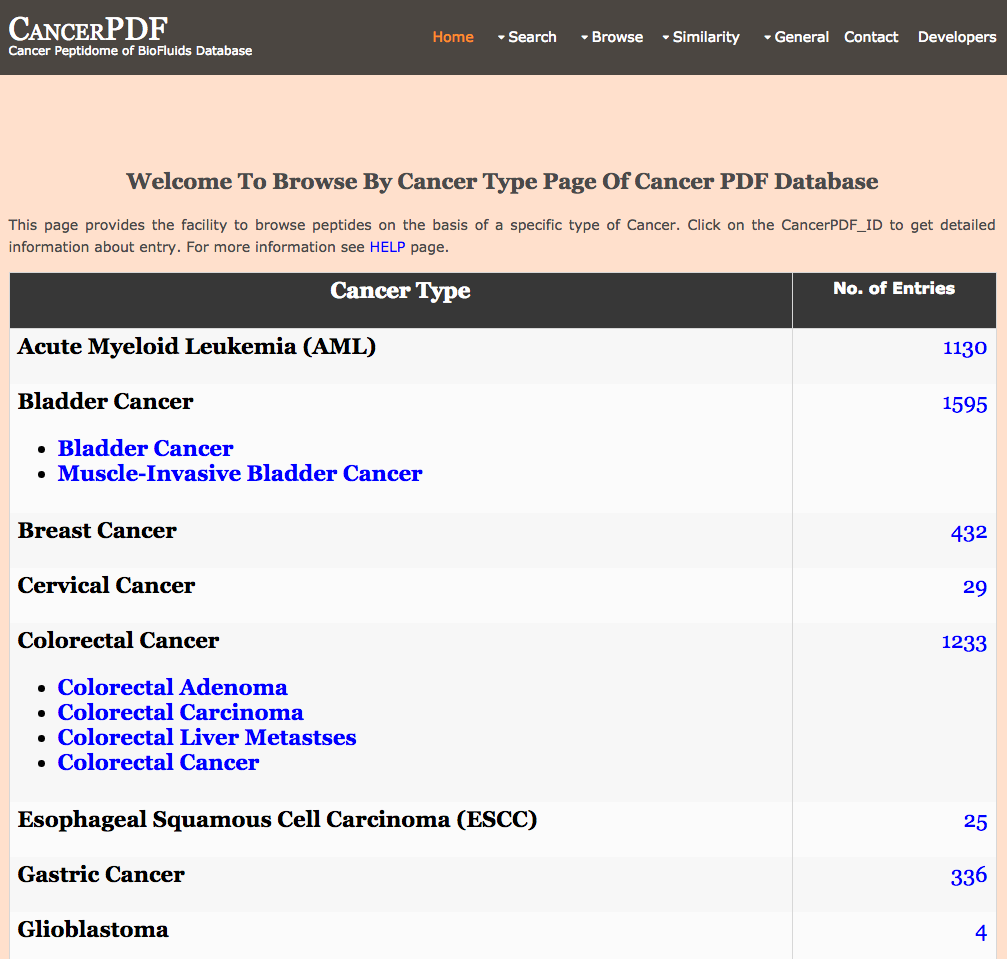
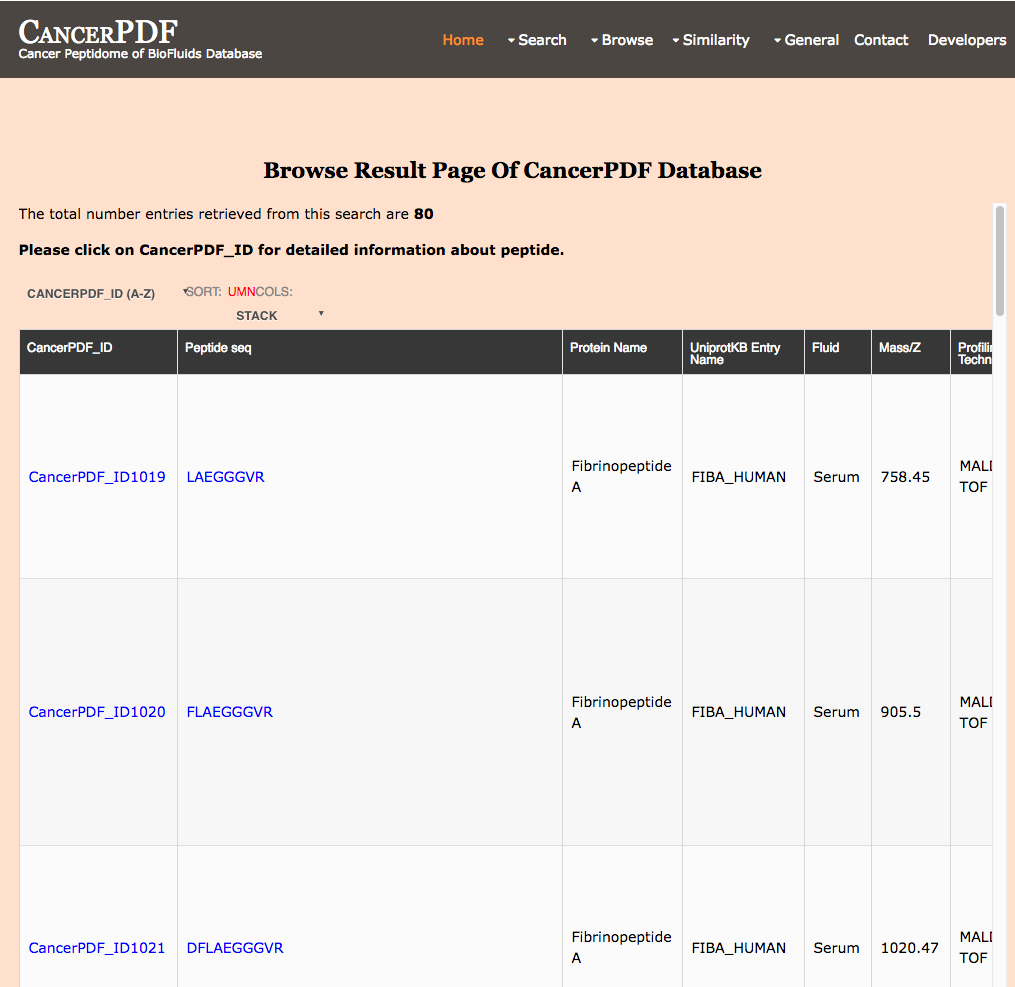
Browse by Cancer Type
This page provides the user to get the list of total number of peptides according to a particular cancer type.
|
|
| Result |
|
|
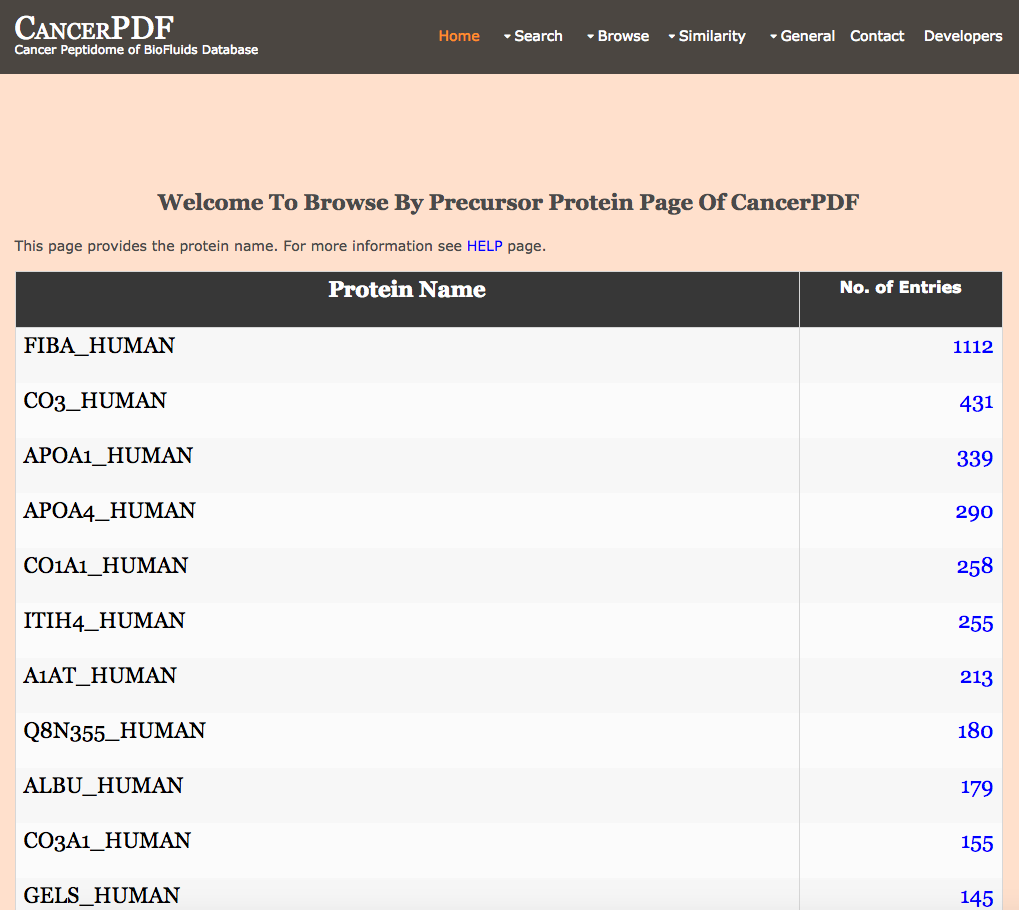
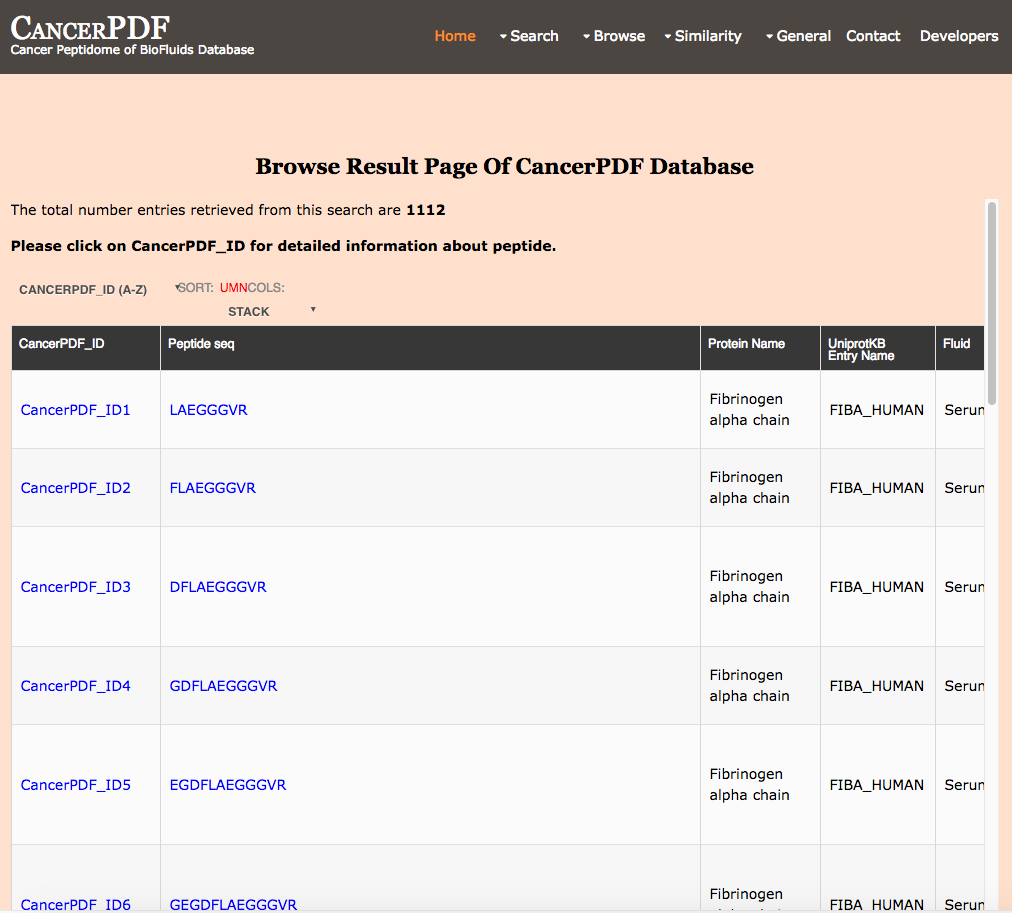
Browse by Precursor Protein
This page provides the user to get the list of all the protein names in the database and clicking.
|
|
| Result |
|
|
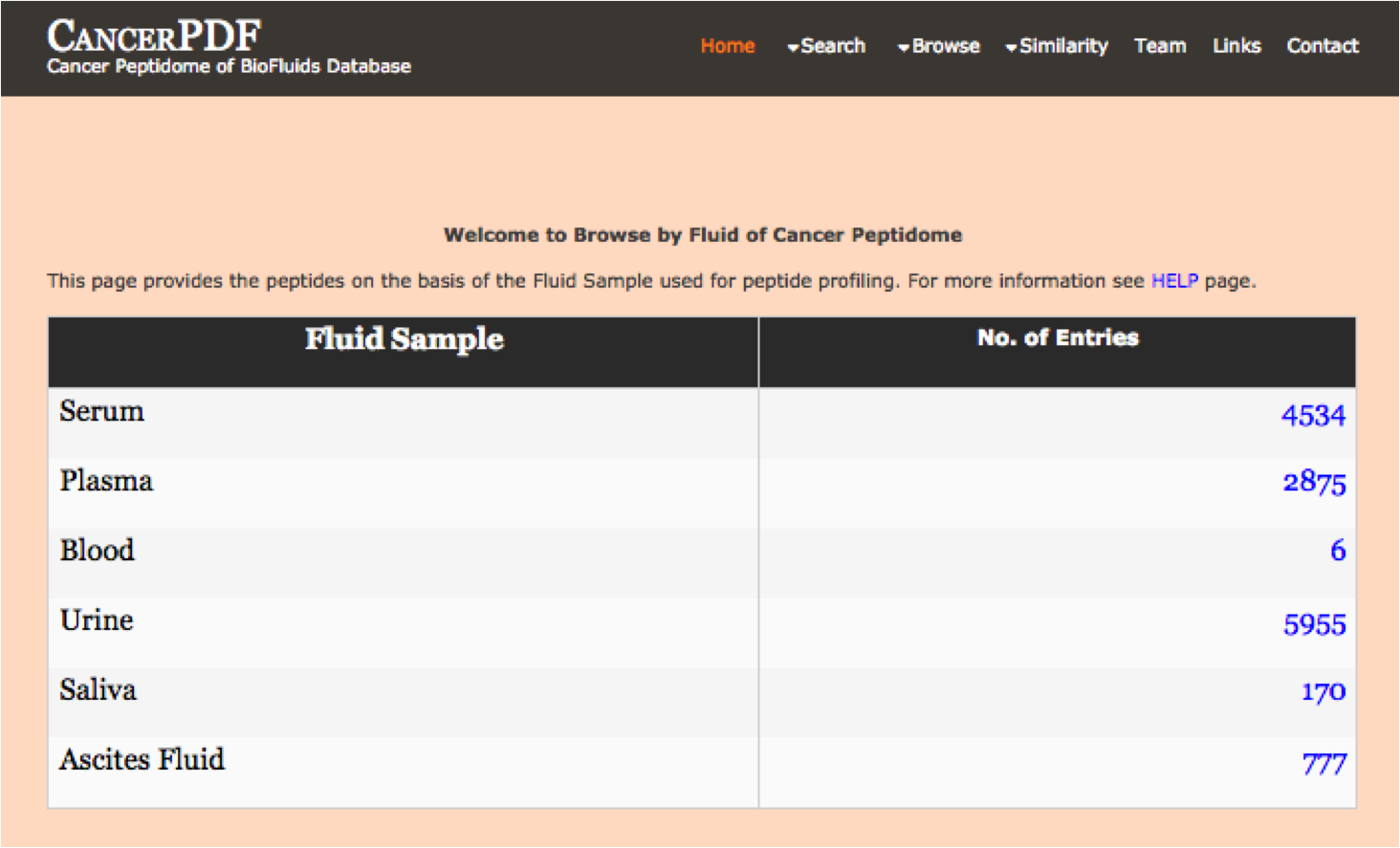
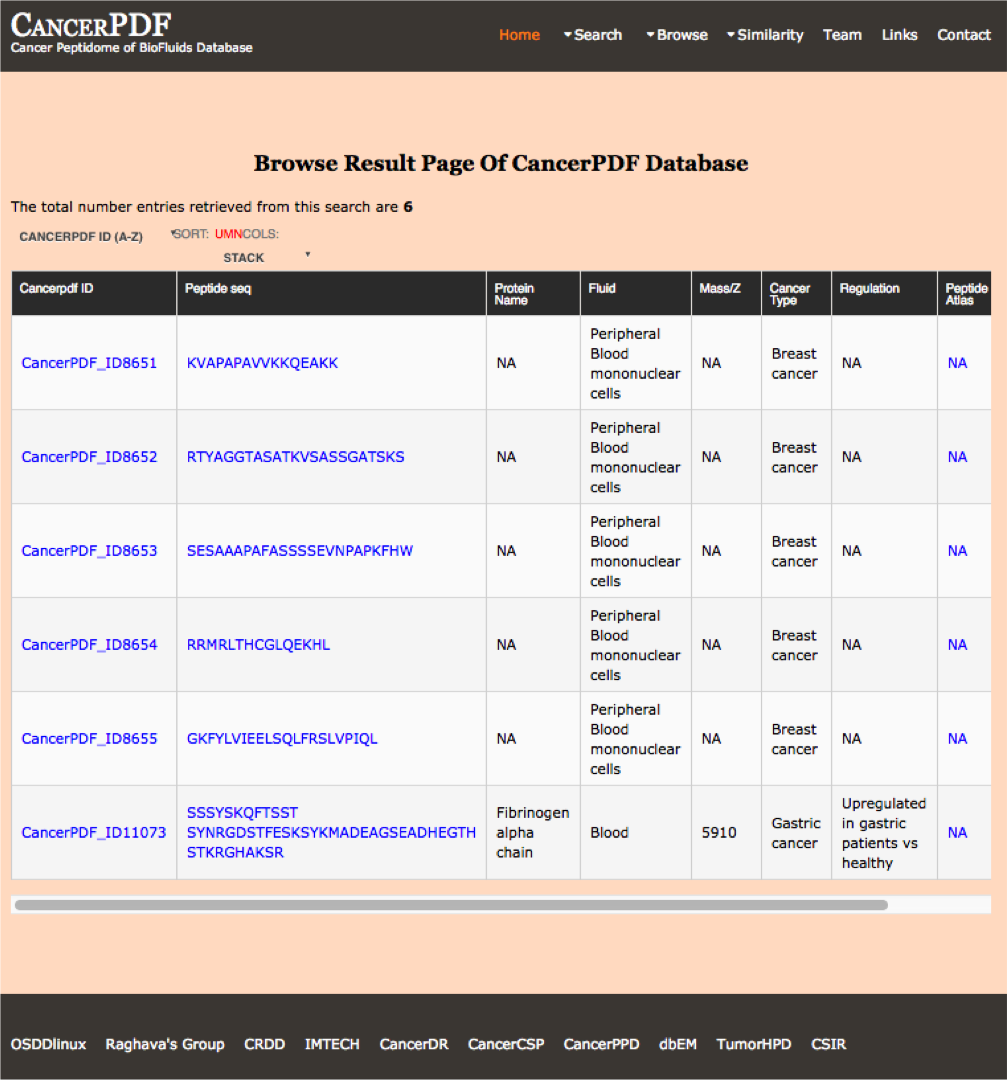
Biofluid
This module displays the peptides present according to the different biofluids in the database. By clicking the different menu user can get number and additional details about the type of peptide selected.
|
|
| Result |
|
|
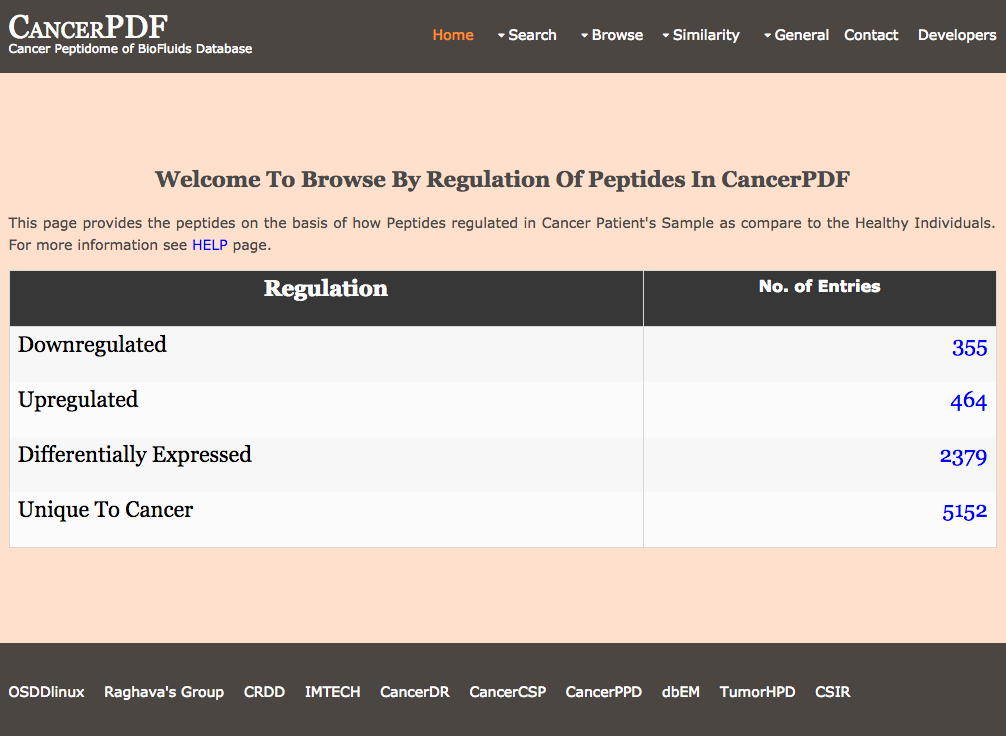
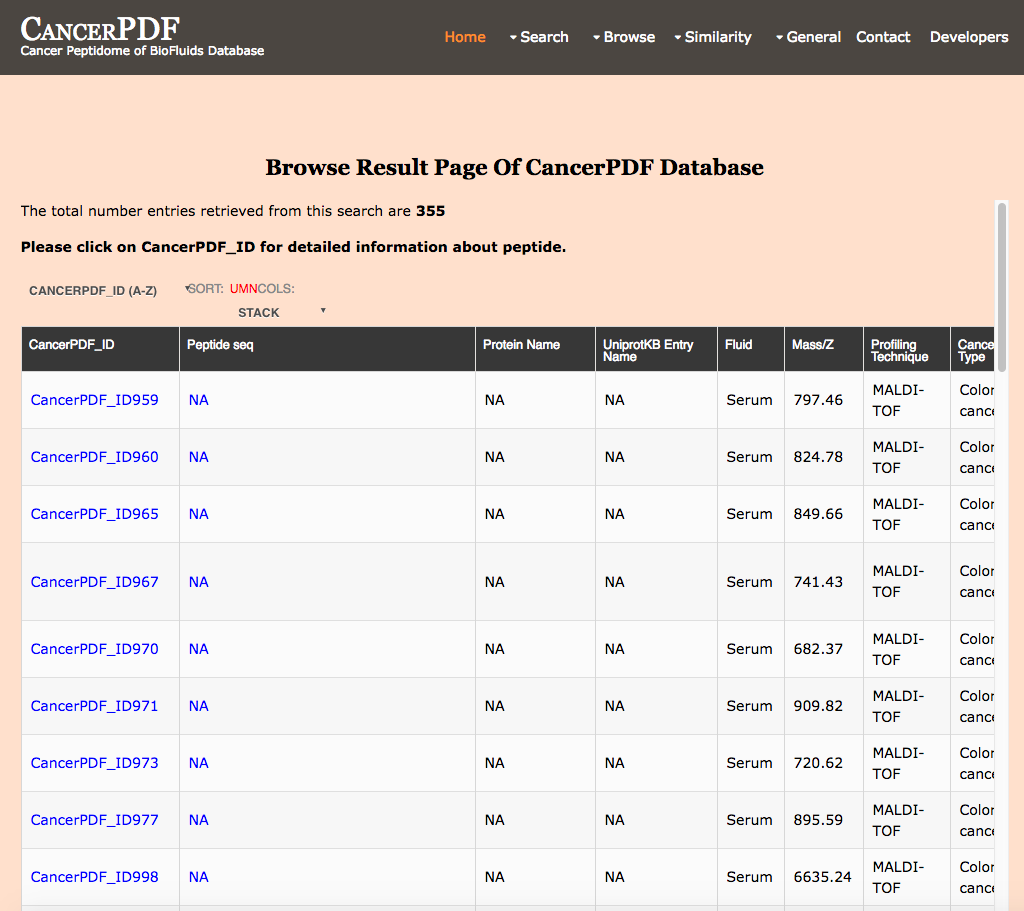
Browse by Regulation
This page provides the user to get the list of total number of Peptides differentially regulated in Cancer Patient's Sample as compare to the Healthy Individuals.
|
|
| Result |
|
|
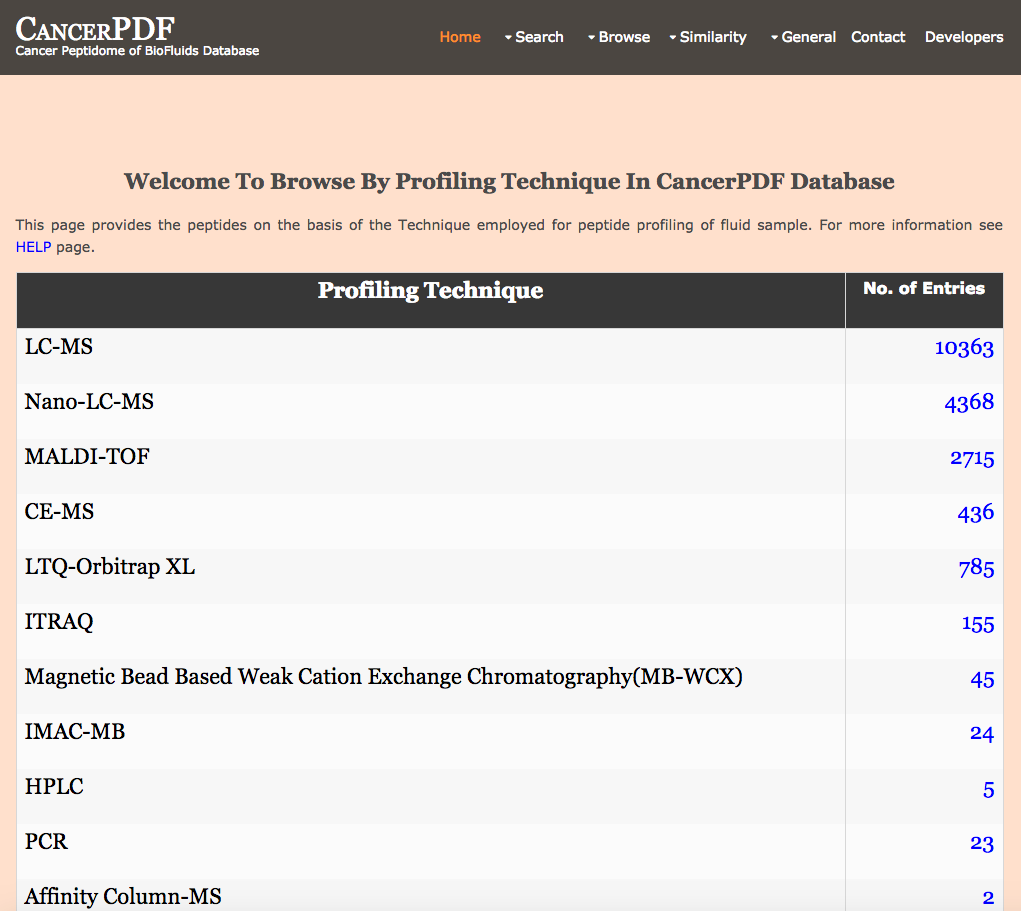
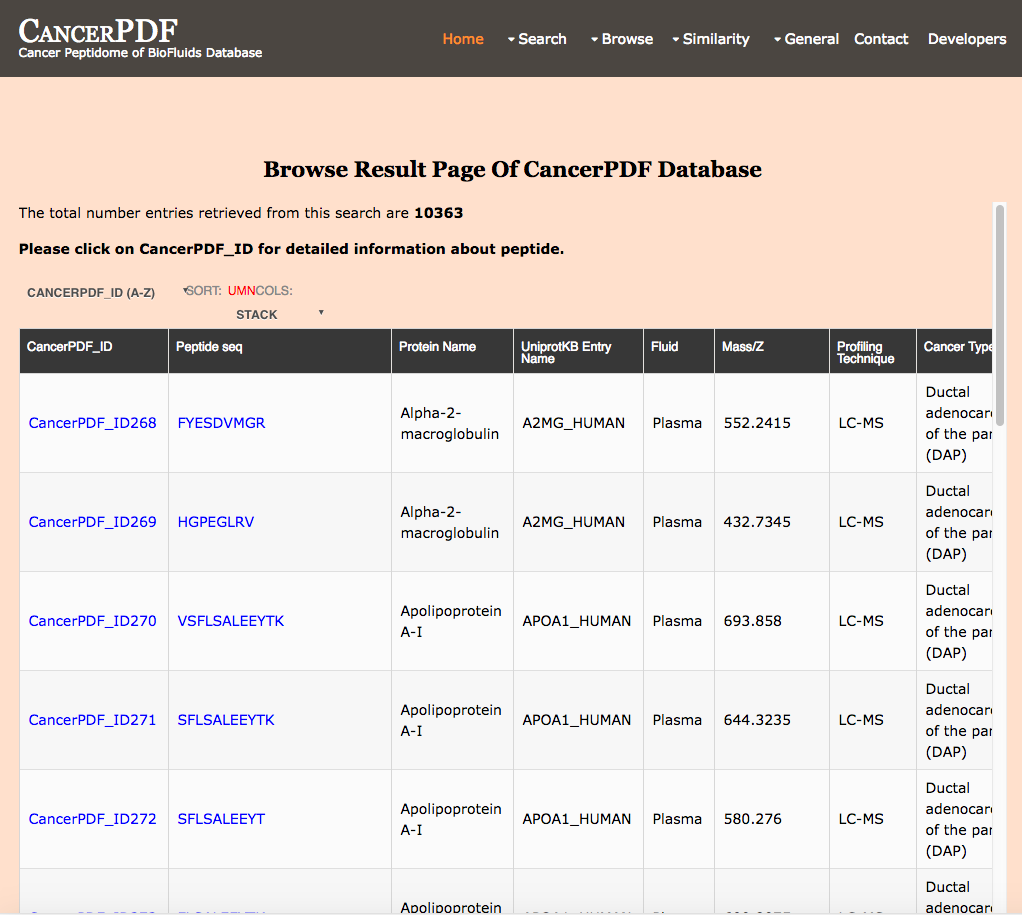
Browse by Profiling Technique
This page provides the user to get the list of total number of peptides on the basis of the Technique employed for peptide profiling of biofluid samples.
|
|
| Result |
|
|
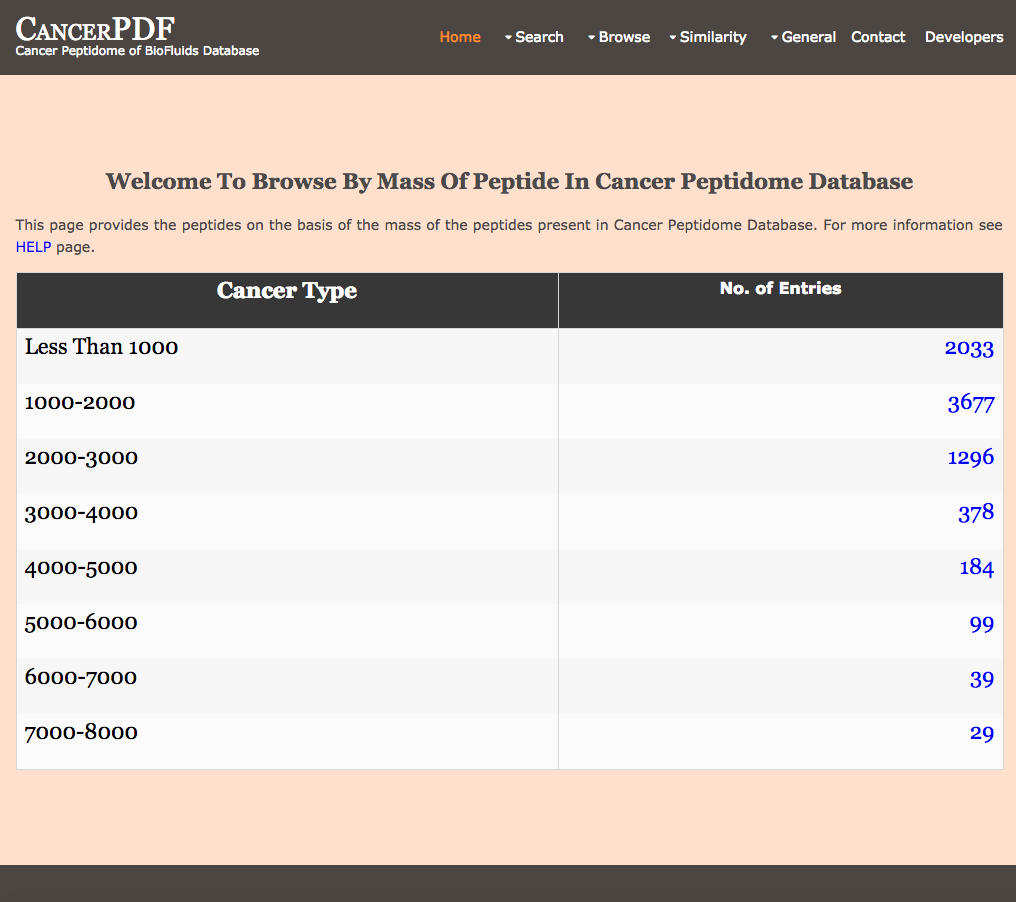
Browse by Mass Range
This page provides the user to get the list of total number of peptides on the basis of the mass of the peptides present in CancerPDF Database.
|
|
| Result |

|
|
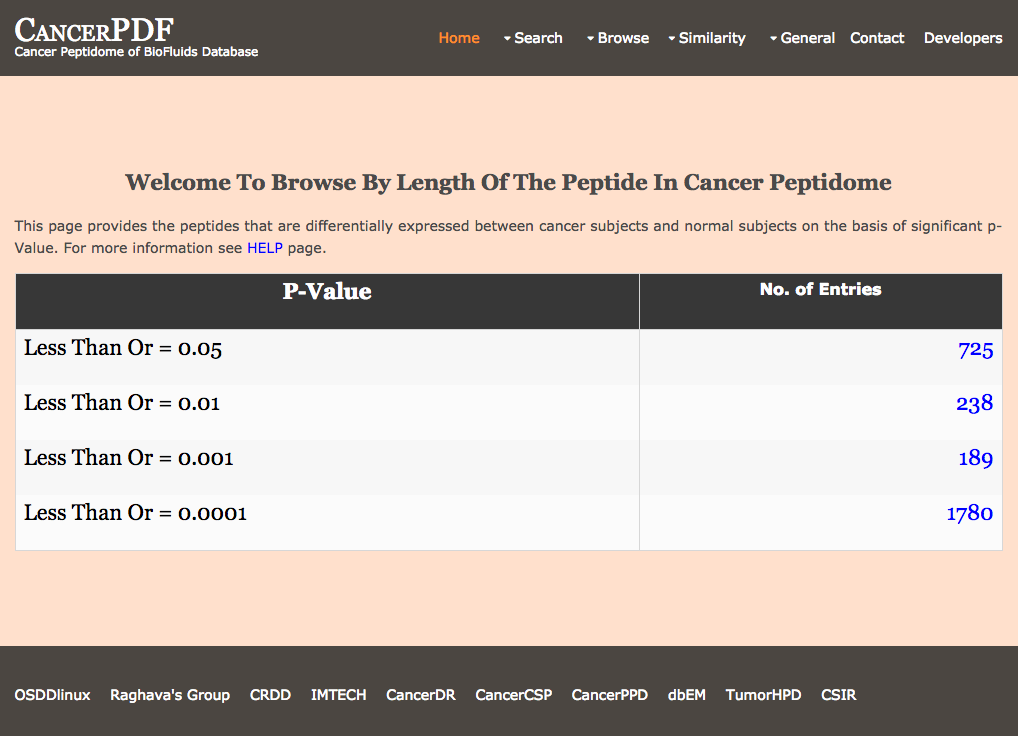
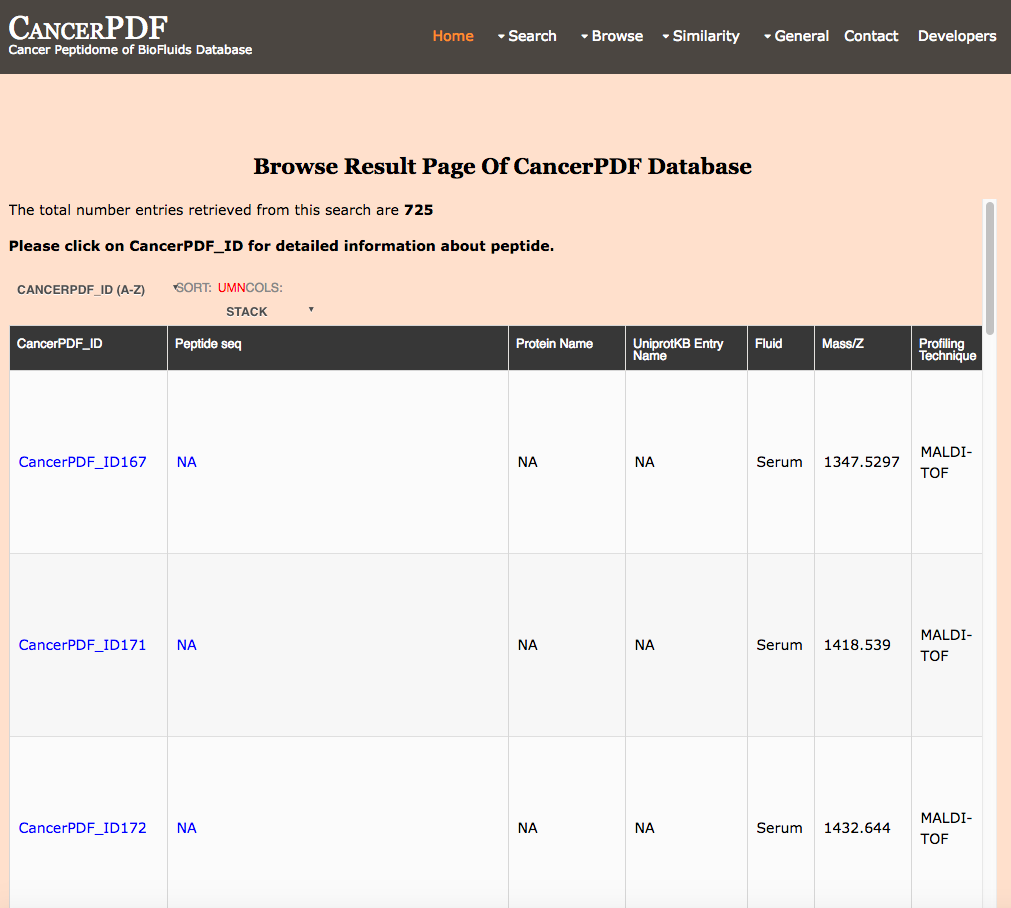
Browse by p-Value
This page provides the user to get the list of total number of peptides that are differentially expressed between cancer subjects and normal subjects on the basis of significant p-value.
|
|
| Result |
|
|
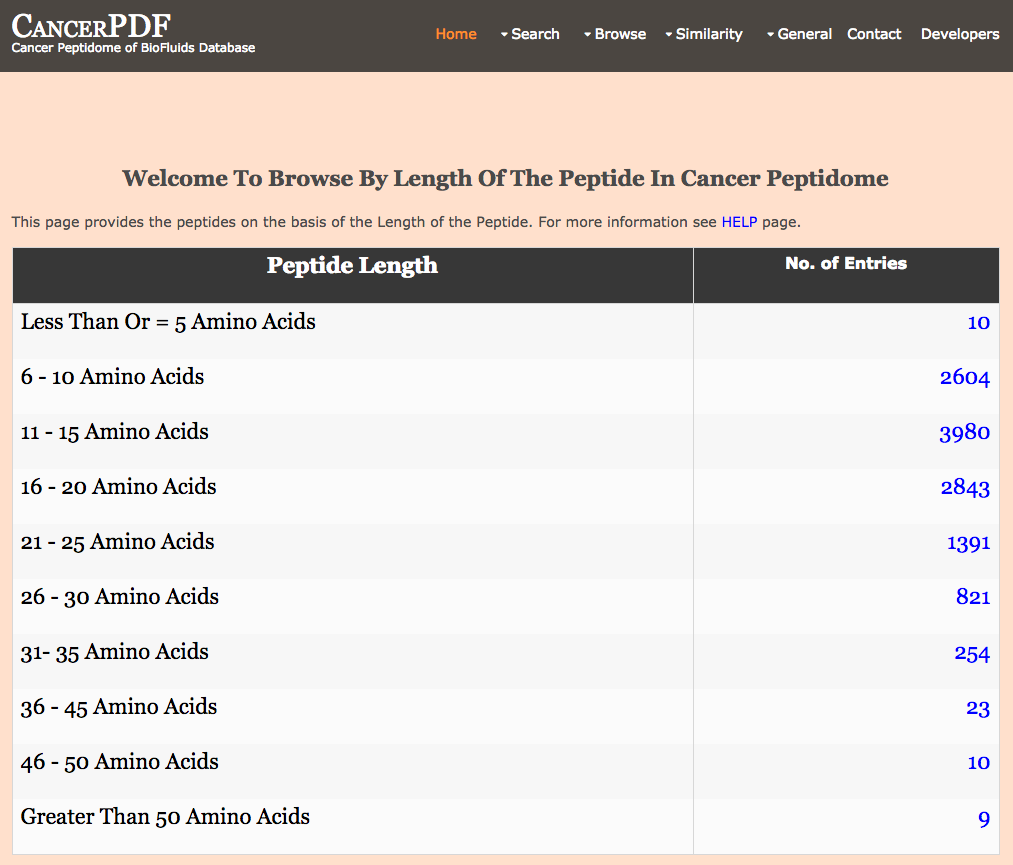
Browse by Peptide Length
This page provides the user to get the list of total number of peptides on the basis of the Length of the Peptide in the cancerpdf database.
|
|
| Result |

|
|
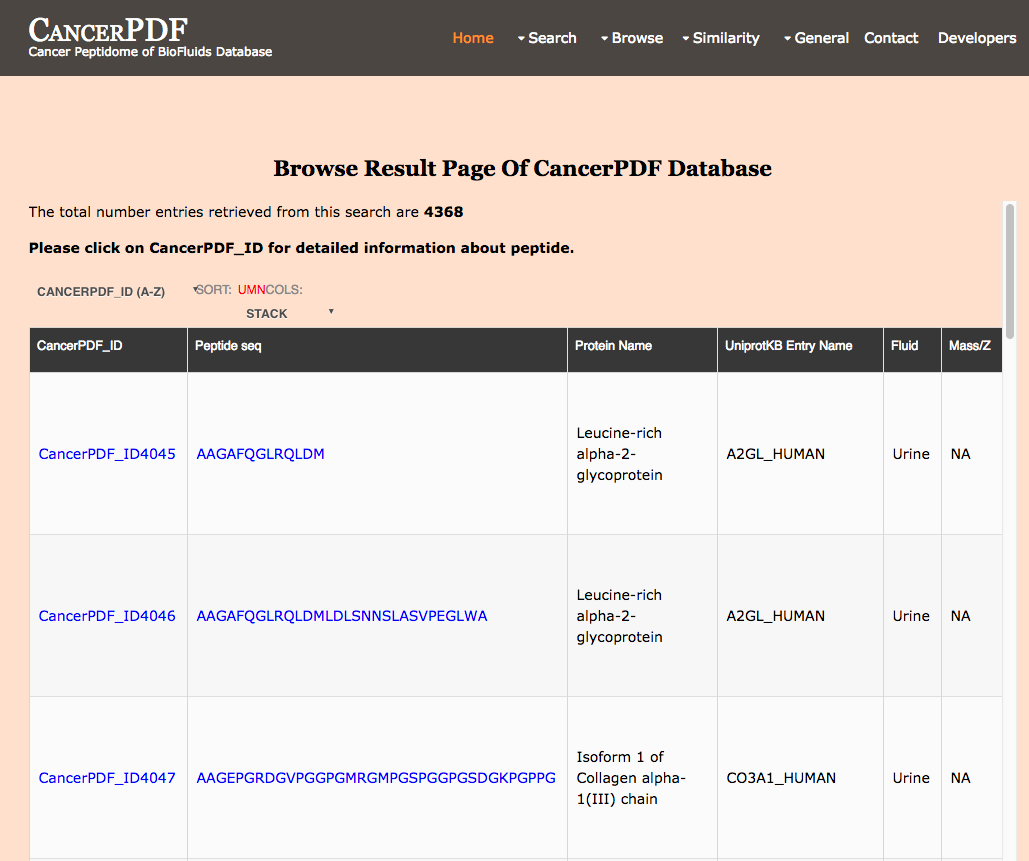
Browse by Pubmed ID
This page provides the user to get the list of total number of peptides by pubmed ID.
|

|
| Result |
|
|
Similarity |
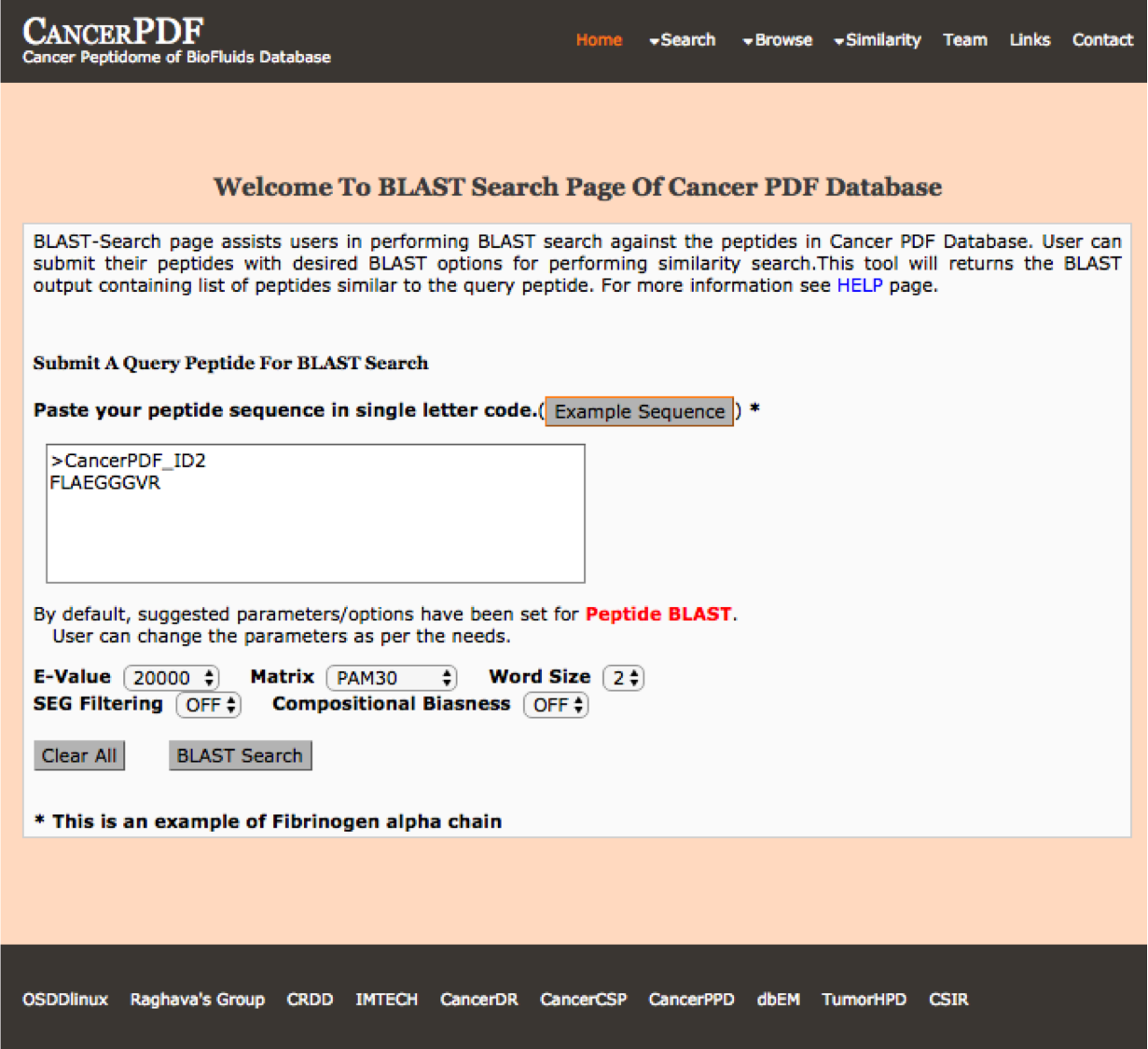
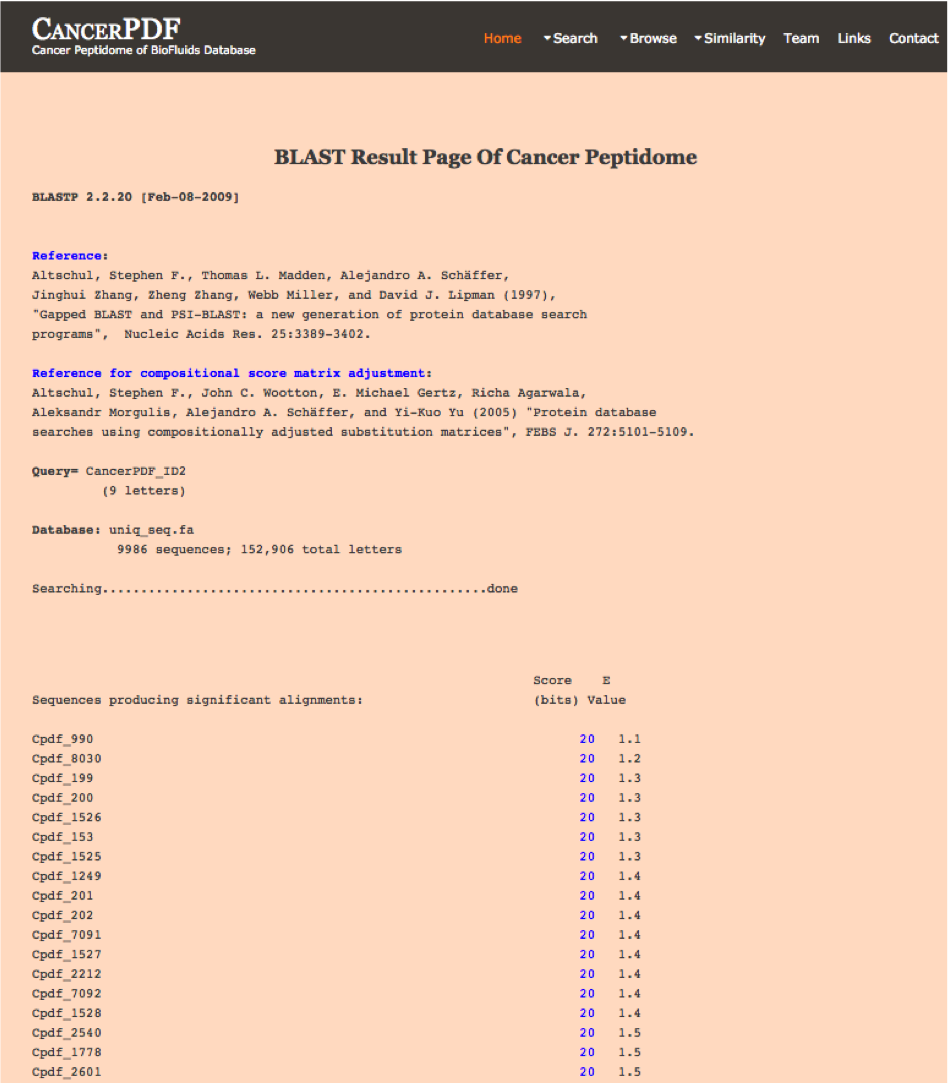
BLAST Search
Blast-Search page assists users in performing BLAST search against the peptides present in CancerPDF. User can submit their peptide with desired BLAST options for performing similarity, server will returns the BLAST output containing list of peptides similar to the query peptide.
|
|
| Result |
|
|
Smith-Waterman
This page allows users to perform similarity search against peptides present in CancerPDF using Smith-Waterman algorithm. User can submit their peptide; server will returns the list of peptides similar to the query peptide.
|
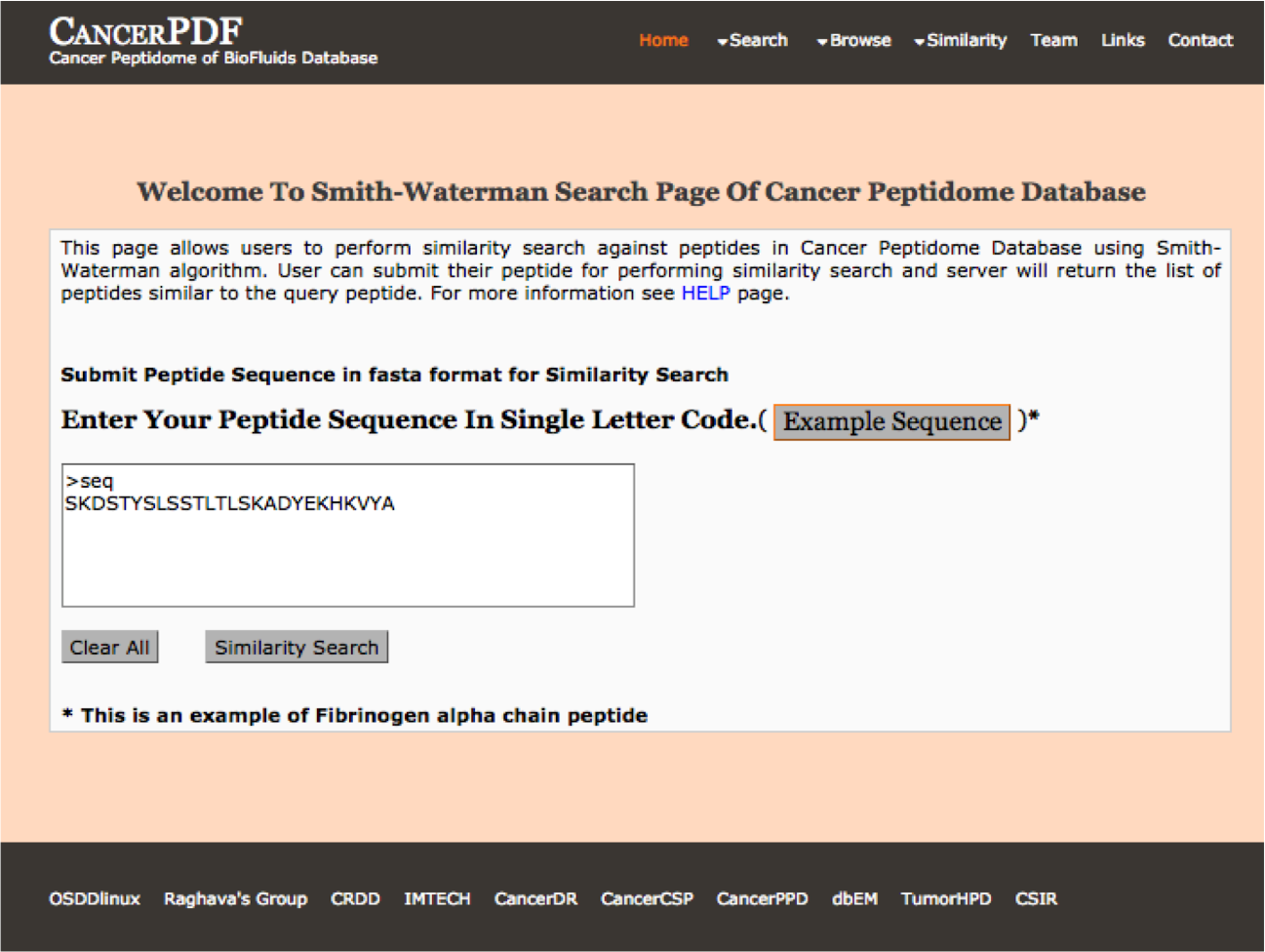
|
| Result |
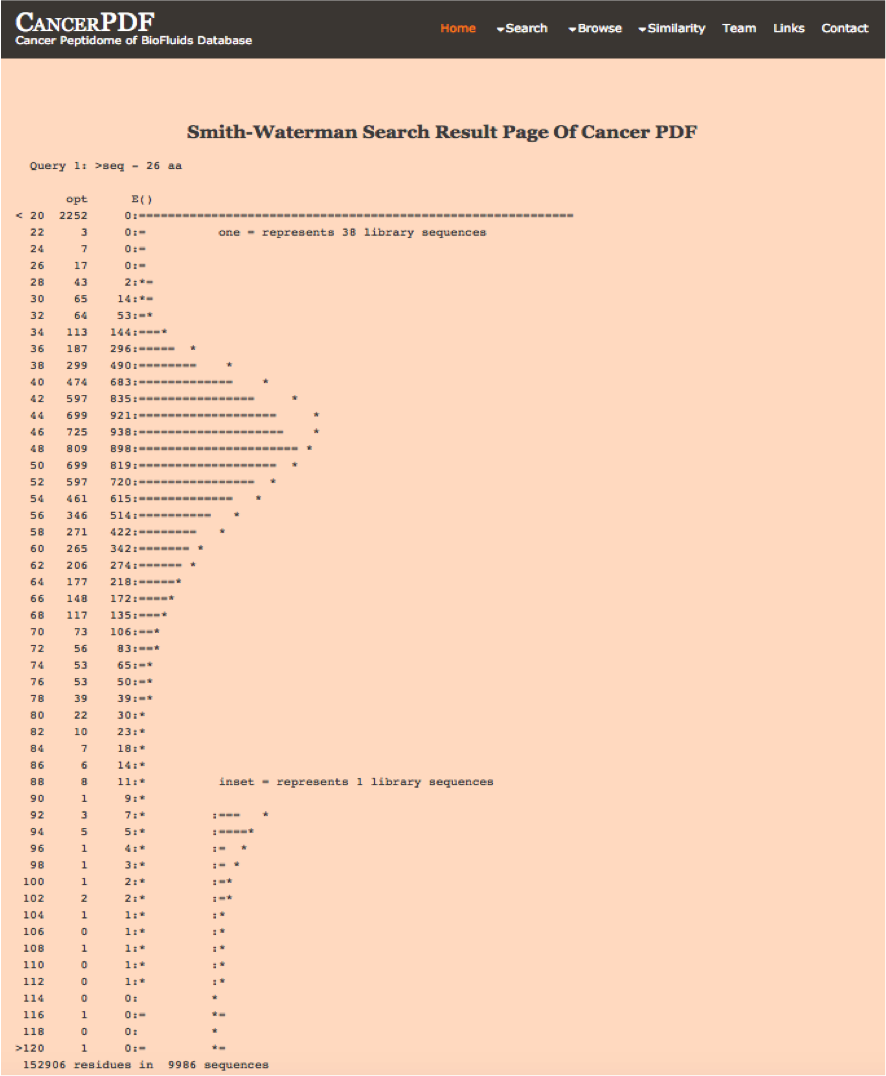
|
|
Multiple Aligment
This page allows users to perform Multiple alignment against peptides present in CancerPDF. User can submit their peptide; server will returns the alignment of peptides.
|
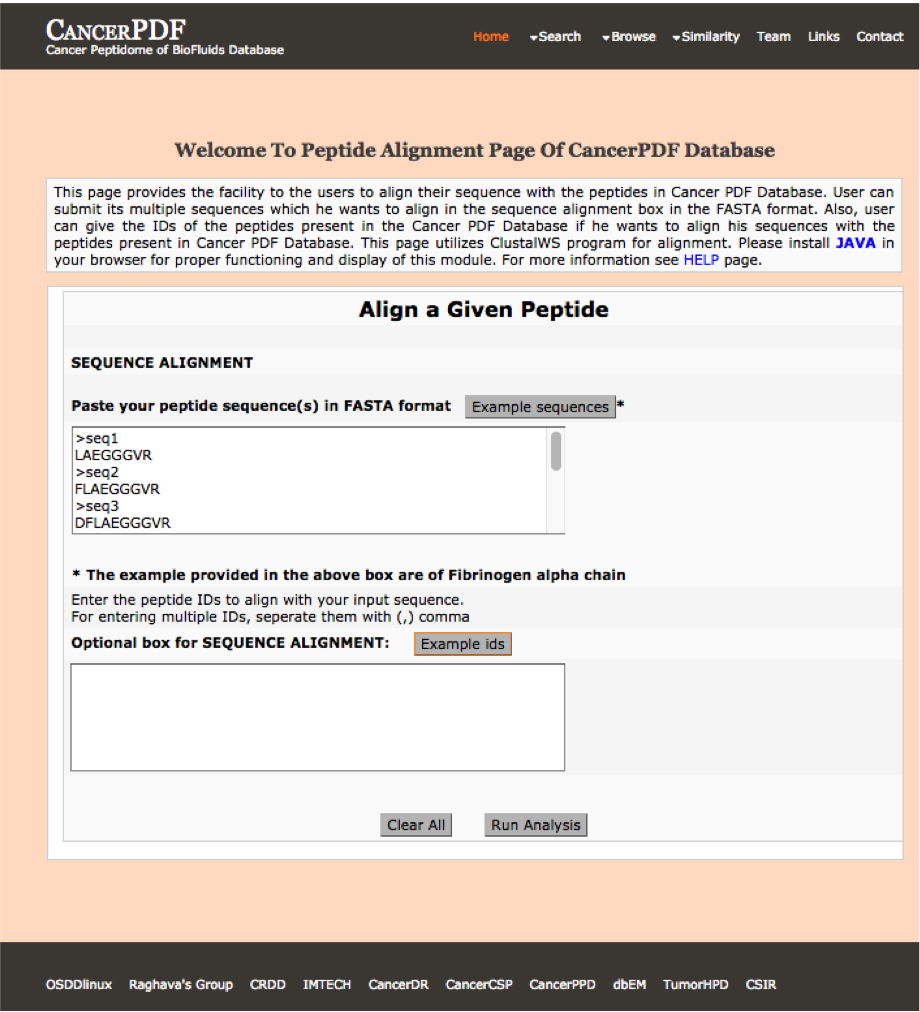
|
Mapping
Peptide Mapping module offers the user to run a sub-search and super-search. In sub-search a given peptide is mapped against all peptides of CancerPDF Database. While super-search returns similar peptides of our database against protein sequence given as query.
|

|

























