Help Page of CPPsite |
| CPPsite is a manually cuarted database of experimentally validated 843 CPPs. Information about CPPs was collected and compiled from literature, patents and public resources. Each entry provides comprehensive information of a peptide that includes peptide name, PubMed ID, peptide sequence, chirality, origin, nature of peptide, sub-cellular localization, uptake efficiency, uptake mechanism, hydrophobicity, amino acid frequency and composition, etc. To make it a user-friendly database, many search tools, browsing options and analysis tools such as BLAST, peptide mapping, etc have been integrated. These tools allow a user to search CPPs based on their amino acid composition, charge, polarity, hydrophobicity, etc. In addition, we have derived various types of information from these peptide sequences that include secondary/tertiary structure, amino acid composition, and physicochemical properties of peptides. |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
CPPs are short (10-30 amino acids) peptides which are usually consist of basic amino acids (arginine and lysine) and have positive net charge on neutral pH. These peptides show no cell type specificity and are able to penetrate the plasma membrane in a receptor independent manner withoutcausing significant membrane damage. In addition, they have enormous capability to translocate \
various conjugated cargos such as proteins, siRNA, liposomes, and nanoparticles inside the cell with high efficiency. Therefore, they are attractive tools for drug delivery.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| On the basis of origin, CPPs can be classified into following three main classes:
(1) Protein derived: This is well known and the largest family of CPPs. These CCPPs are part of natural proteins and also known as protein transduction domain (PTD). e.g. Tat and penetratin.
(3) Synthetic: These are designed peptides. e.g. oligoarginine and model amphipathic peptide (MAP). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
The uptake mechanism of CPPs depends on many factors such as size (cargo), temperature, cell types and nature of CPPs. Despite the tremendous research on CPPs, the internalization mechanismof CPPs is an unresolved issue to date. Two uptake mechanisms have been proposed so far that include (i) endocytosis and (ii) Non- endocytic pathways (direct translocation). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| (1) Endocytosis: Endocytosis is a process of macromolecule internalization that is energy-dependent. It consists of mainly two pathways including phagocytosis for uptake of large particles and pinocytosis for fluids and solutes uptake. Phagocytosis is restricted to specialized c
ells like macrophages and leucocytes, but pinocytosis occurs in all cells. Pinocytosis can occurthrough at least four different endocytosis pathways; clathrin mediated, caveolin mediated, cla\
thrin and caveolin independent endocytosis and macropinocytosis. All four processes have been implicated as pathways for internalization of different CPPs with different types of cargo. Endocytosis is characterized by vesicle formation. (2) Non-endocytic pathways: Direct penetration is also known as energy-independent pathway. Different models/ mechanisms have been described for direct penetration of CPPs, that includes (i) inverted micelle formation (ii) pore formation (iii) carpet-like model and (iv) membrane thinning model. All these models follow a 3-step process of internalization: (a) membrane interaction, (b) membrane permeation and (c) release of CPP into the cytosol | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
SEARCH | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
KEYWORD SEARCH This is a very simple and easy search option. User can search with single word for sequence (e.g. CSNIDARAC), motif (e.g. WYY), target tumor (e.g. Breast cancer), target cell (e.g. Endothelial cell), cell lines (e.g. MDA-MB) used in the experiments. Results will be displayed according to the selected fields to be dispalyed. e.g. sequence, research article, PMID etc. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
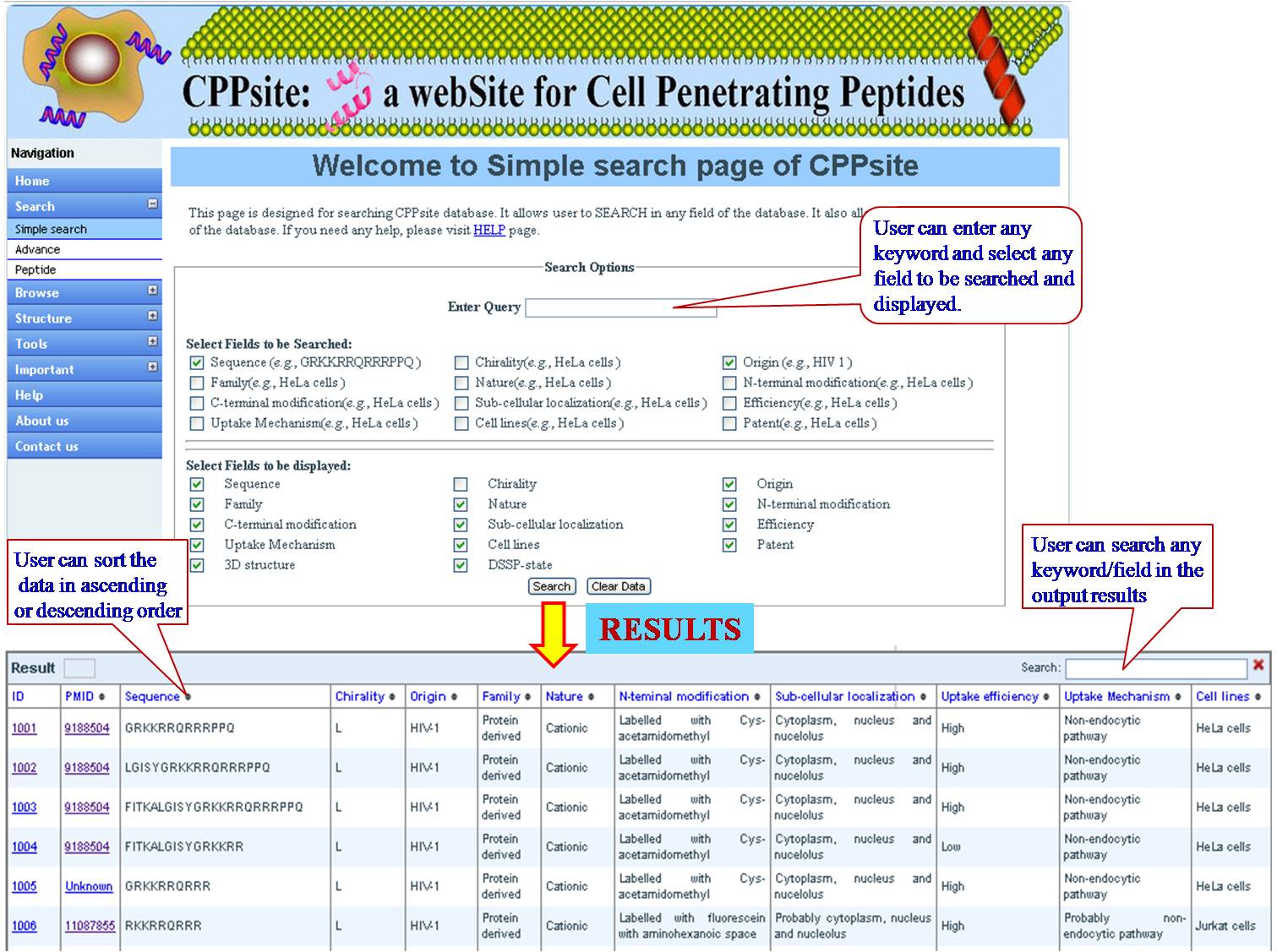 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
ADVANCED SEARCH This search has multiple options which give very refined result for the user. Here user has been provided with different search options Field name Match option (equal =, less <, greater > ). Value column is provided to add input from user e.g. ID, year, tumor related word etc. and then user can apply conditions(AND & OR). 1. If user is giving ID in the field name and to match it with year e.g. 2011, the user will opt for (equal=), result page will display all the ID with year 2011. 2. If condition AND is applied then another search field will be opened, user can put value like lung, then results with year 2011 containing lung in the field selected will be displayed. 3. If condition OR with the above input, the user will get all 2011 research articles and all of them will have word lung. 4. Match option LIKE, if user is opting for LIKE option then value must be put in between two % symbols e.g. % blood%. Here user will get result searched for blood in the given field option, if the user is putting % in front then result will be displayed where the word blood is in the beginning and if the % is put after word like blood %, then user will get result where blood word is in the end of the field which is choosen. *Note: While submitting, the advanced search for various conditons, the last search box in the conditions must be blank. By pressing search, desired result will be displayed. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
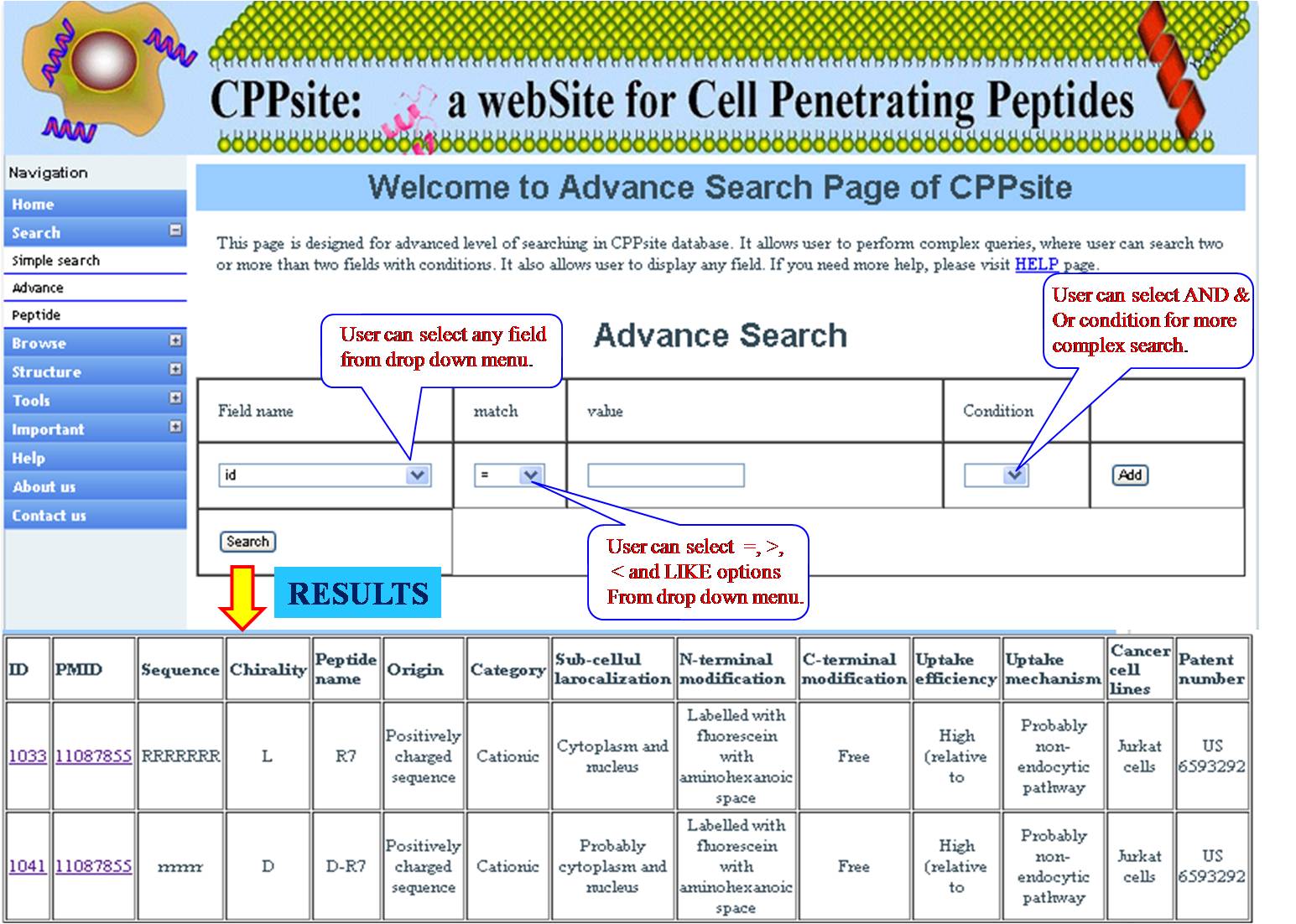 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
PEPTIDE SEARCH User can search the query sequence against whole database. Search result will provide user either with exactly identical peptide or matching peptides with the given query sequence. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
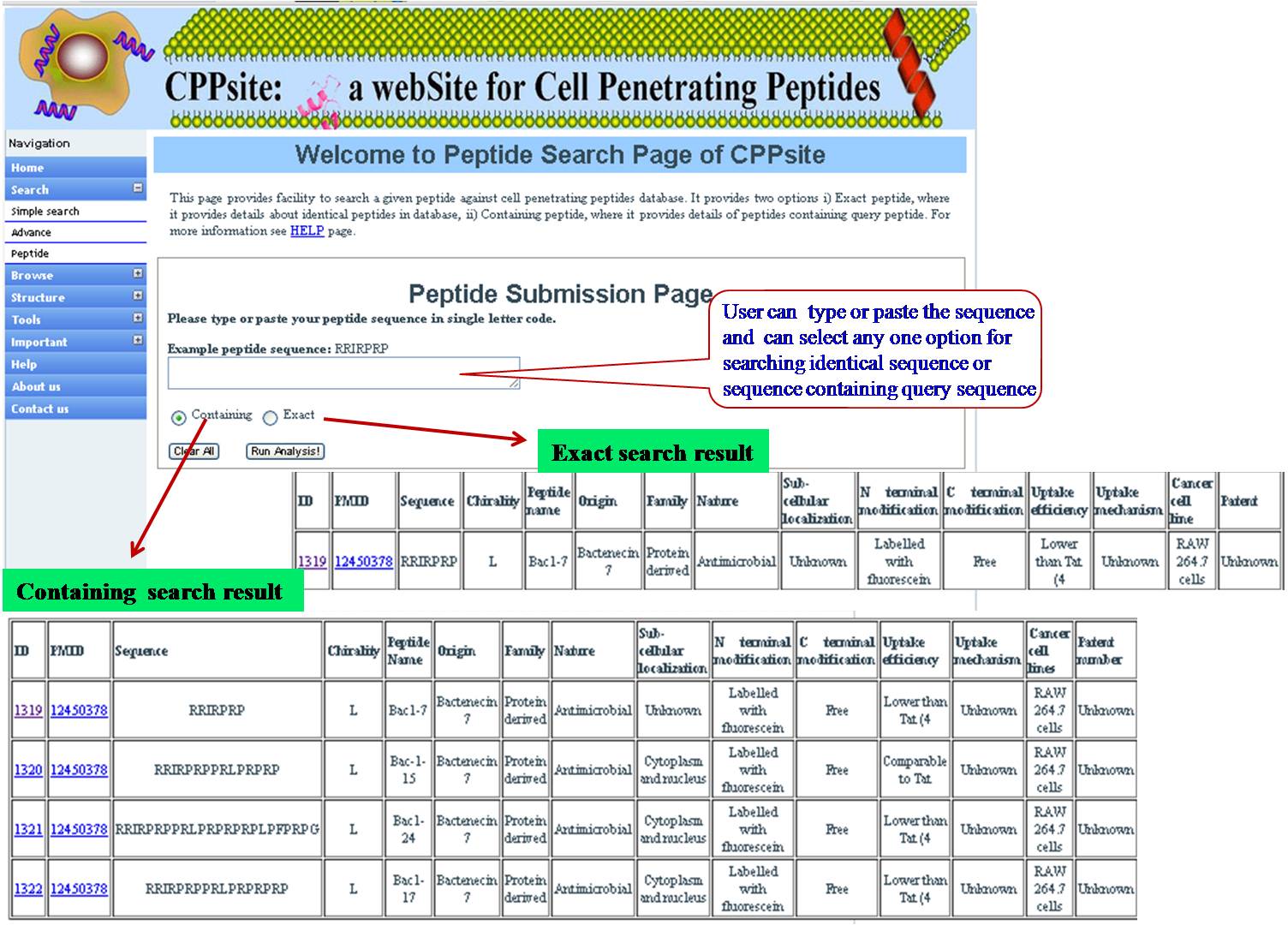 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
BROWSE | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
MAJOR FIELDS This interface allows user to browse database on the following four major fields (1) Target tumor (2) Cell line (3) year of publication (4) Target cell type. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
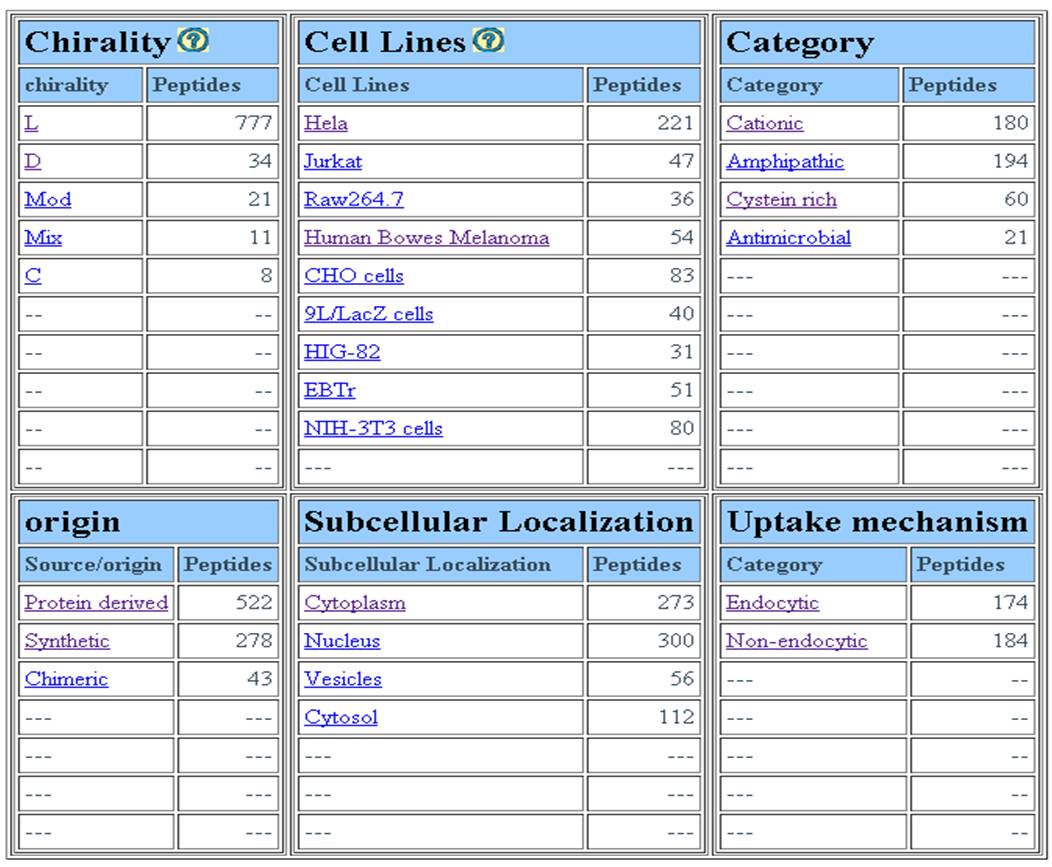 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
AA FREQUENCY AND AA COMPOSITION User can access the database for the amino acid frequency with range between (0-100) e.g. if the user enters lysine value 3 for minimum and 23 for maximum, the user will get list of all the sequences in the database with lysine value between 3 and 23. If the user is putting values for another amino acid along with lysine, e.g. Glycine 4 min and 18 max, then user will get list of sequences which has been sorted for lysine with desired value of glycine. Same procedure is followed for amino acid composition, Physical property frequency and composition. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
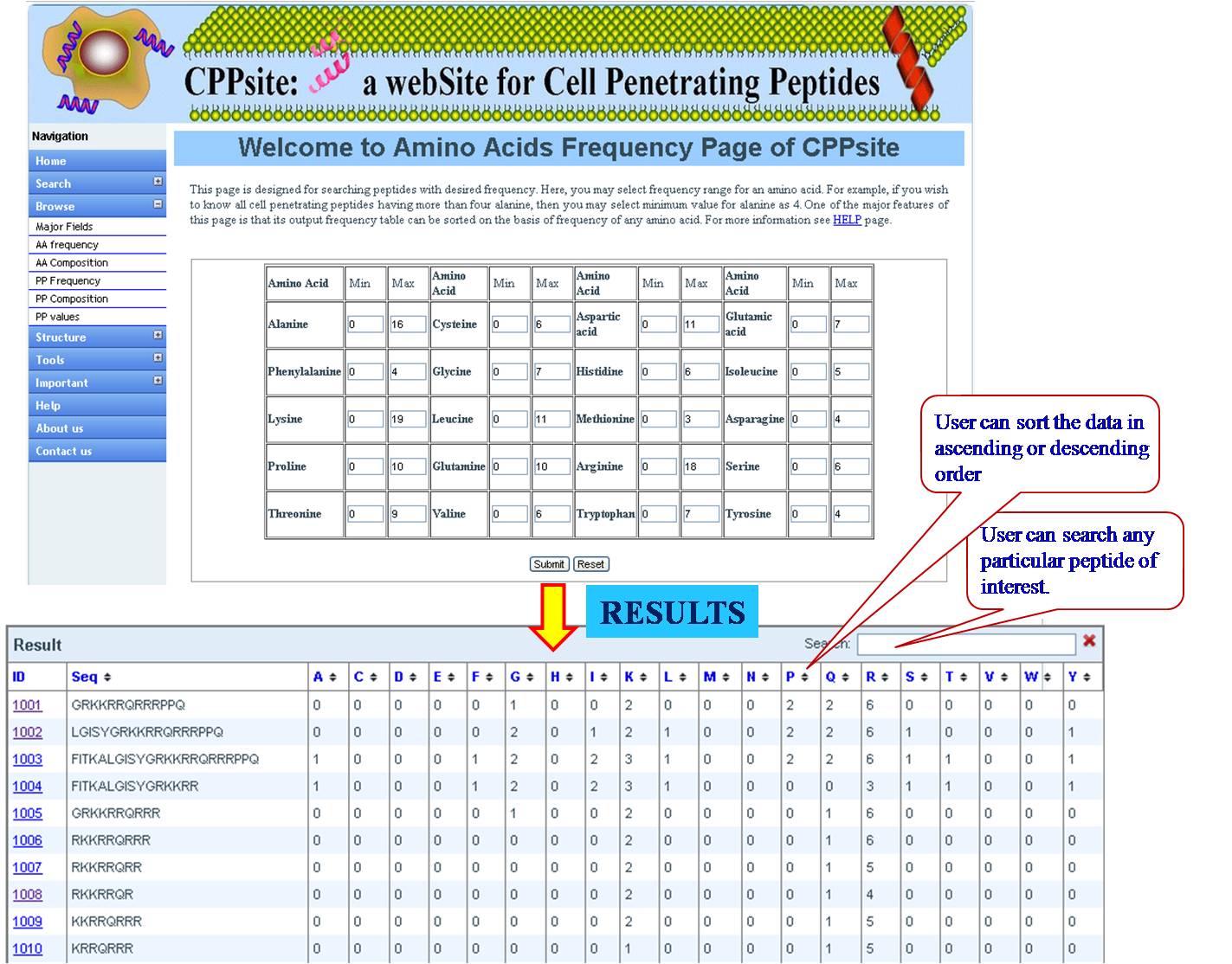 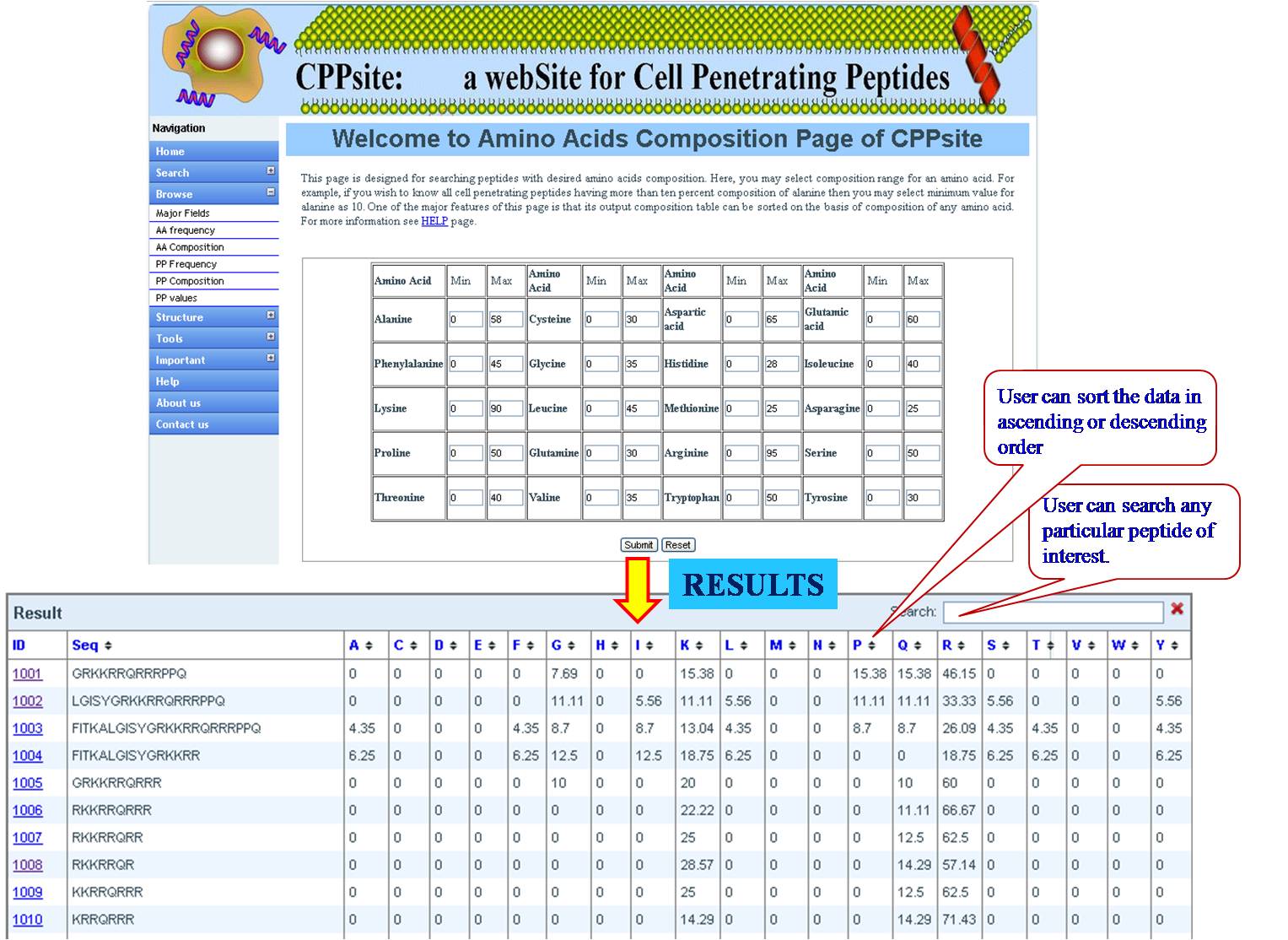 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
PHYSICOCHEMICAL PROPERTY (PP) FREQUENCY AND COMPOSITION User can extract tumor homing peptides that have desired frequency of certain types of residues like, positive charge, negative charge, polar residues. By default we have provided minimum and maximum values for our dataset. User can set the desired range for that property and peptides will be sorted e.g. if we give a range for aromatic property between (2-8), all peptides between this range will be listed in the output table. User can give combination of properties like aromatic property and positive charge, within the desired range. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
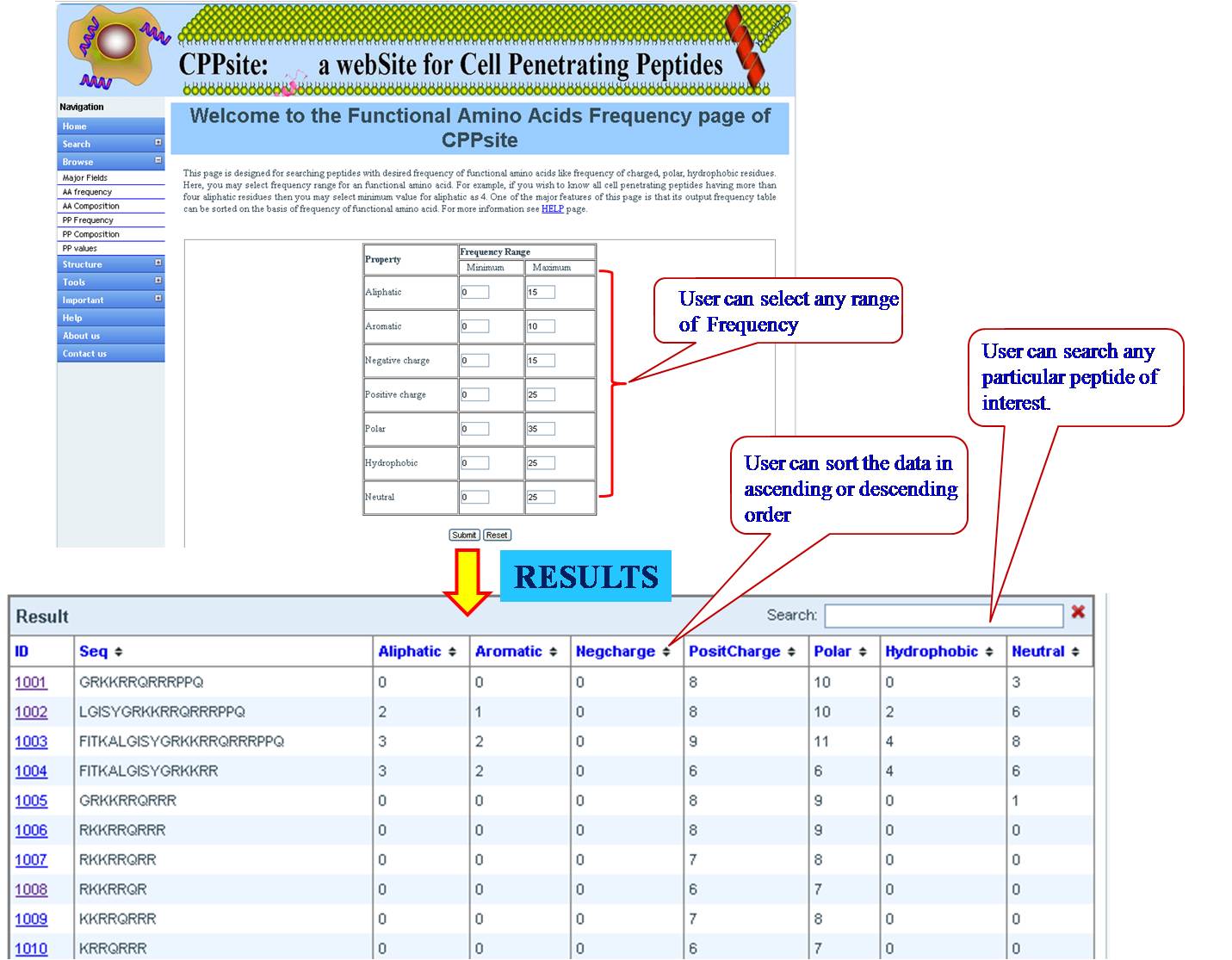 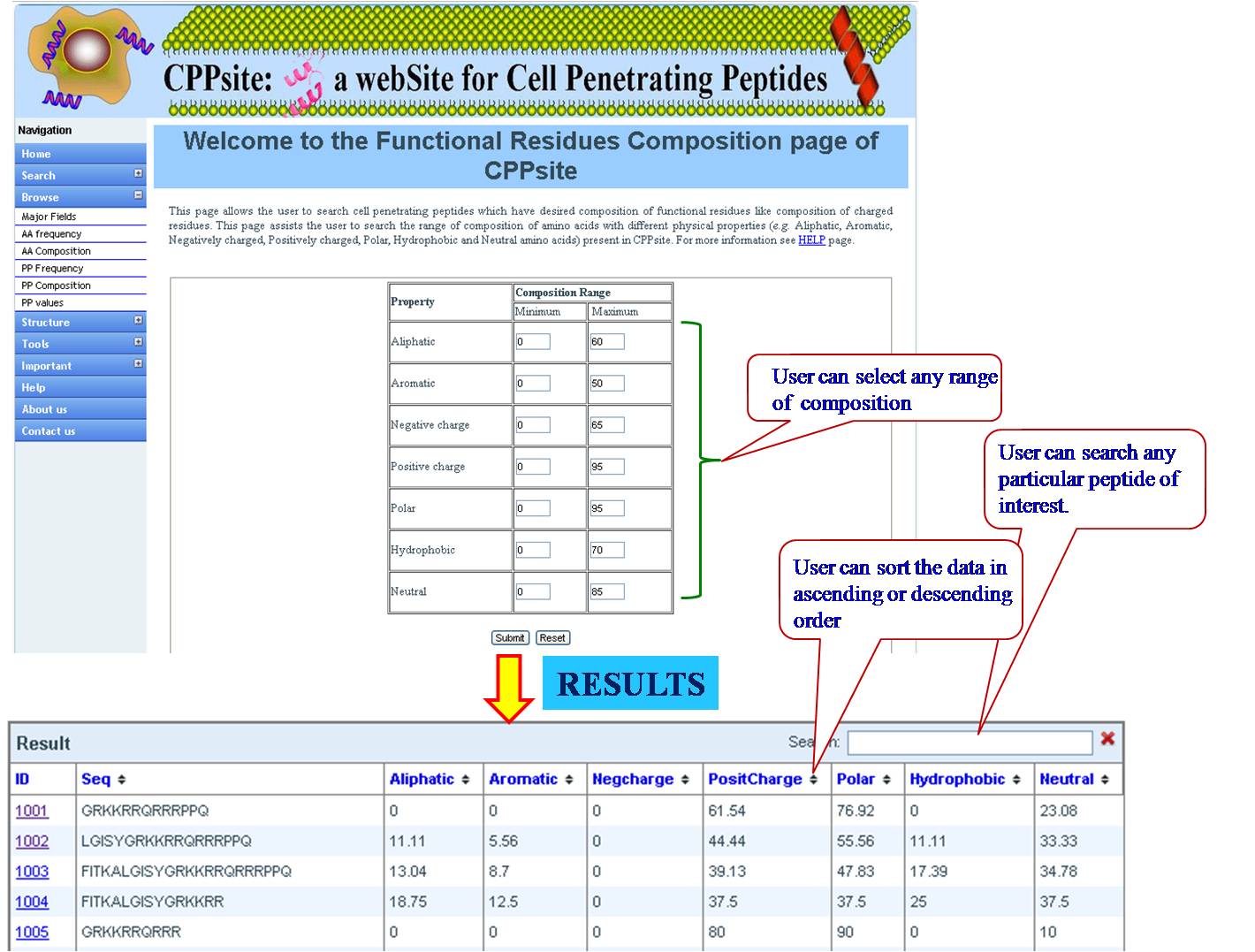 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
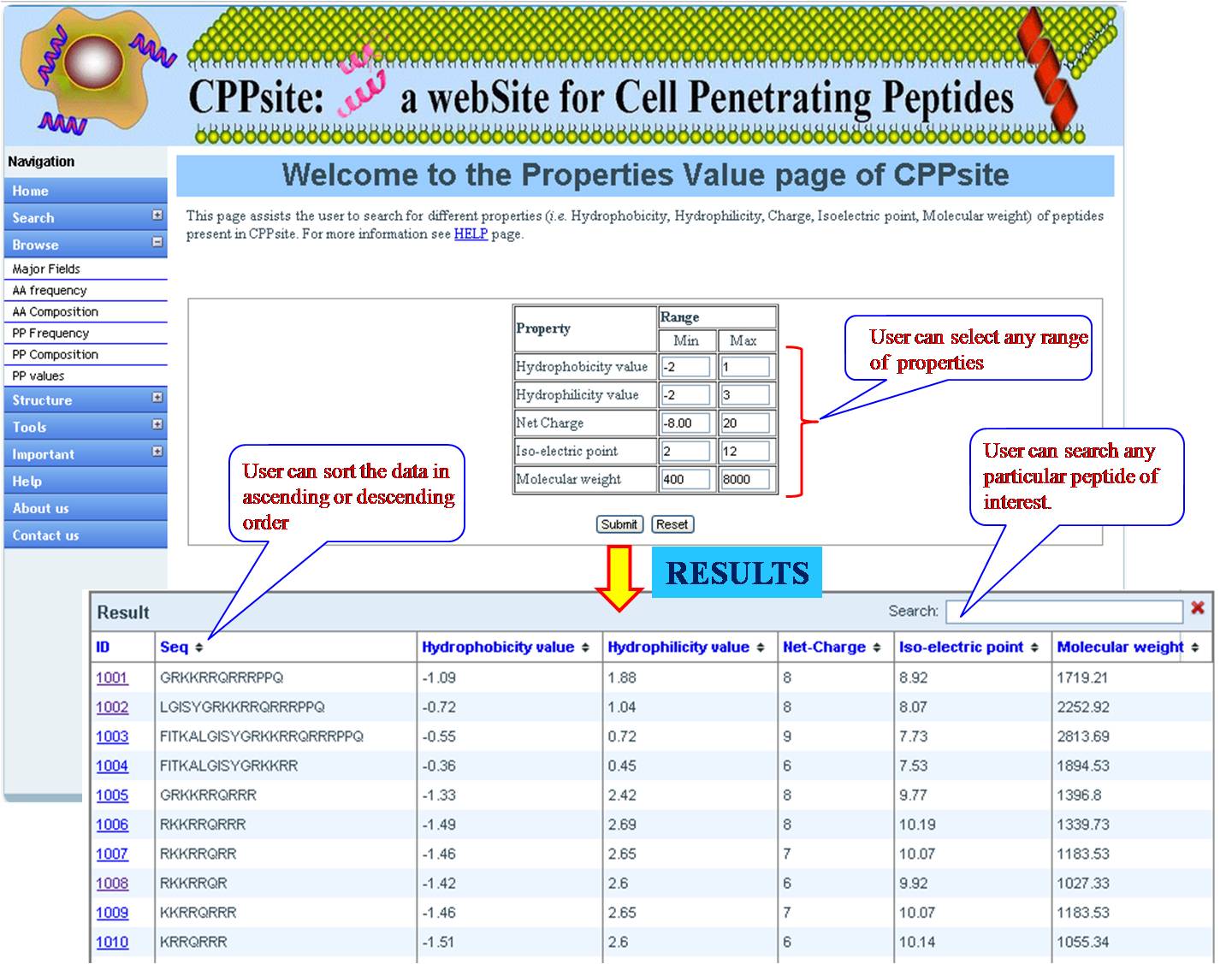
PHYSICOCHEMICAL PROPERTY VALUES This option provides user with list of tumor homing peptides with specified range (according to user) for Hydrophobicity value, Hydrophilicity value, Net charge, Isoelectric point and molecular weight. User has to fill the minimum and maximum value, and will get desired peptides in the above category. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
STRUCTURE | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
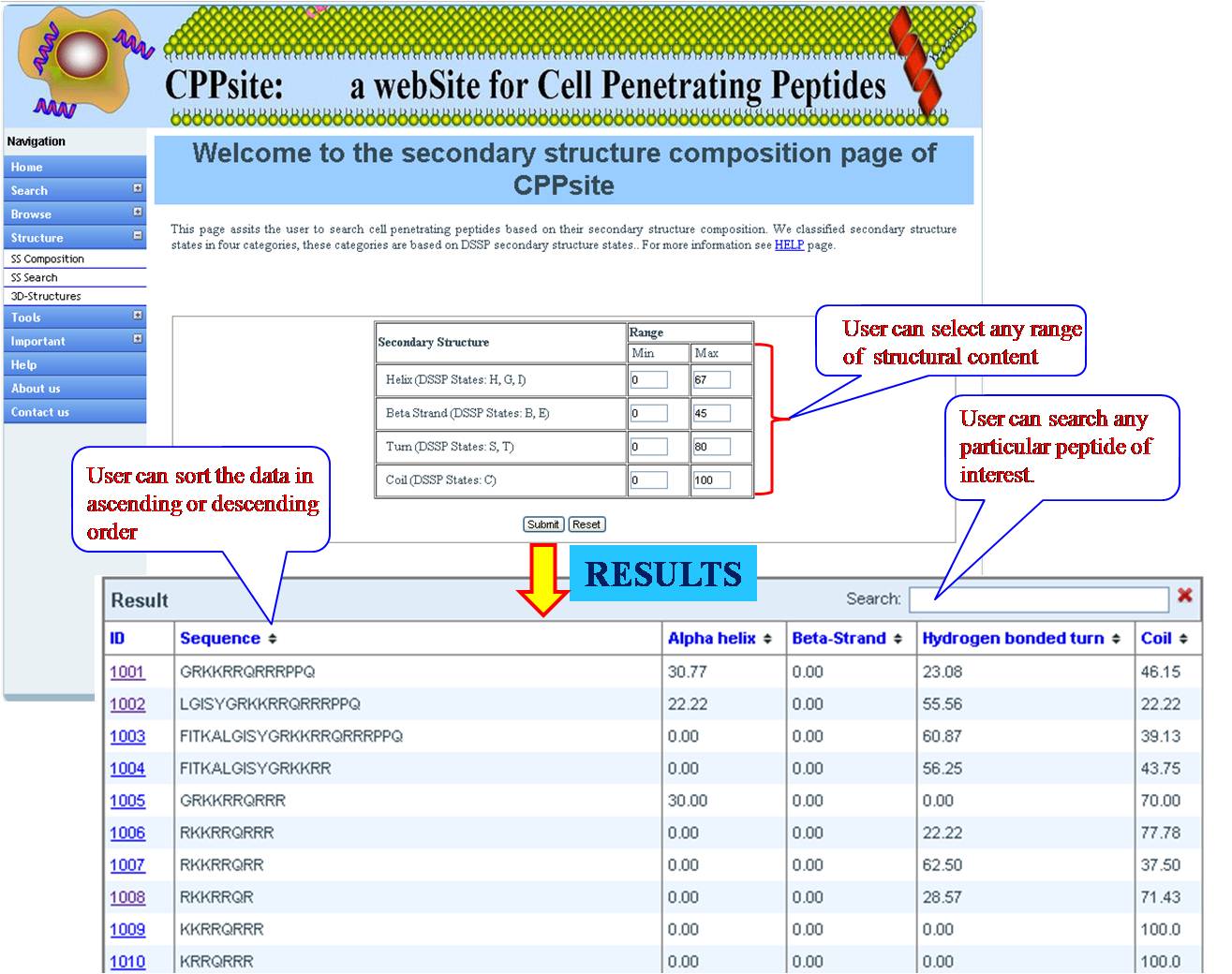
SS COMPOSITION User can get list of tumor homing peptides on the basis of percent composition of four different secondary structural states (Helix -H, Betasheet-E, Turn-T, Coil -C) with their desired range of minimum and maximum value. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
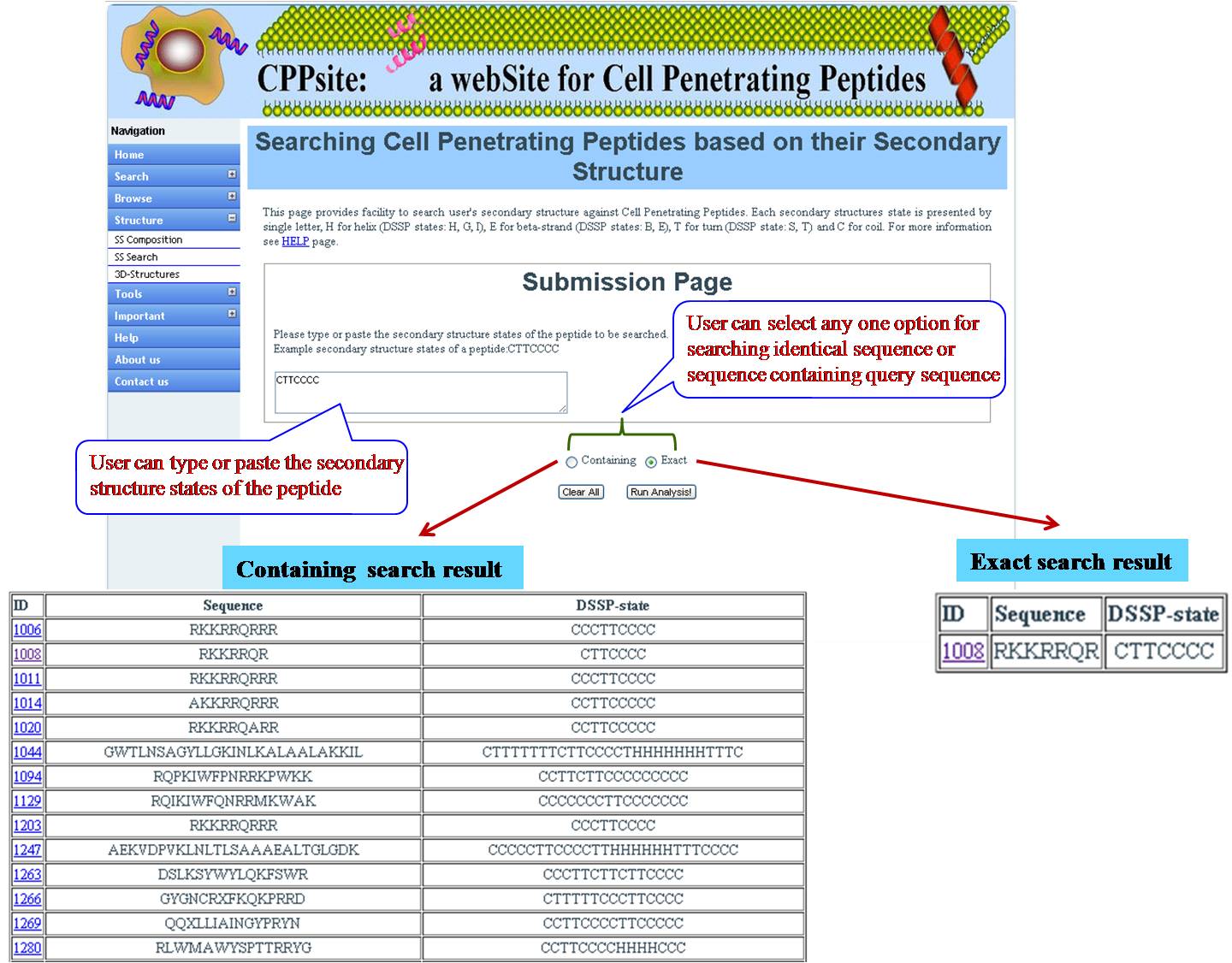
SS SEARCH In this option user has to submit the peptide sequence from the database and user will get predicted structural state. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
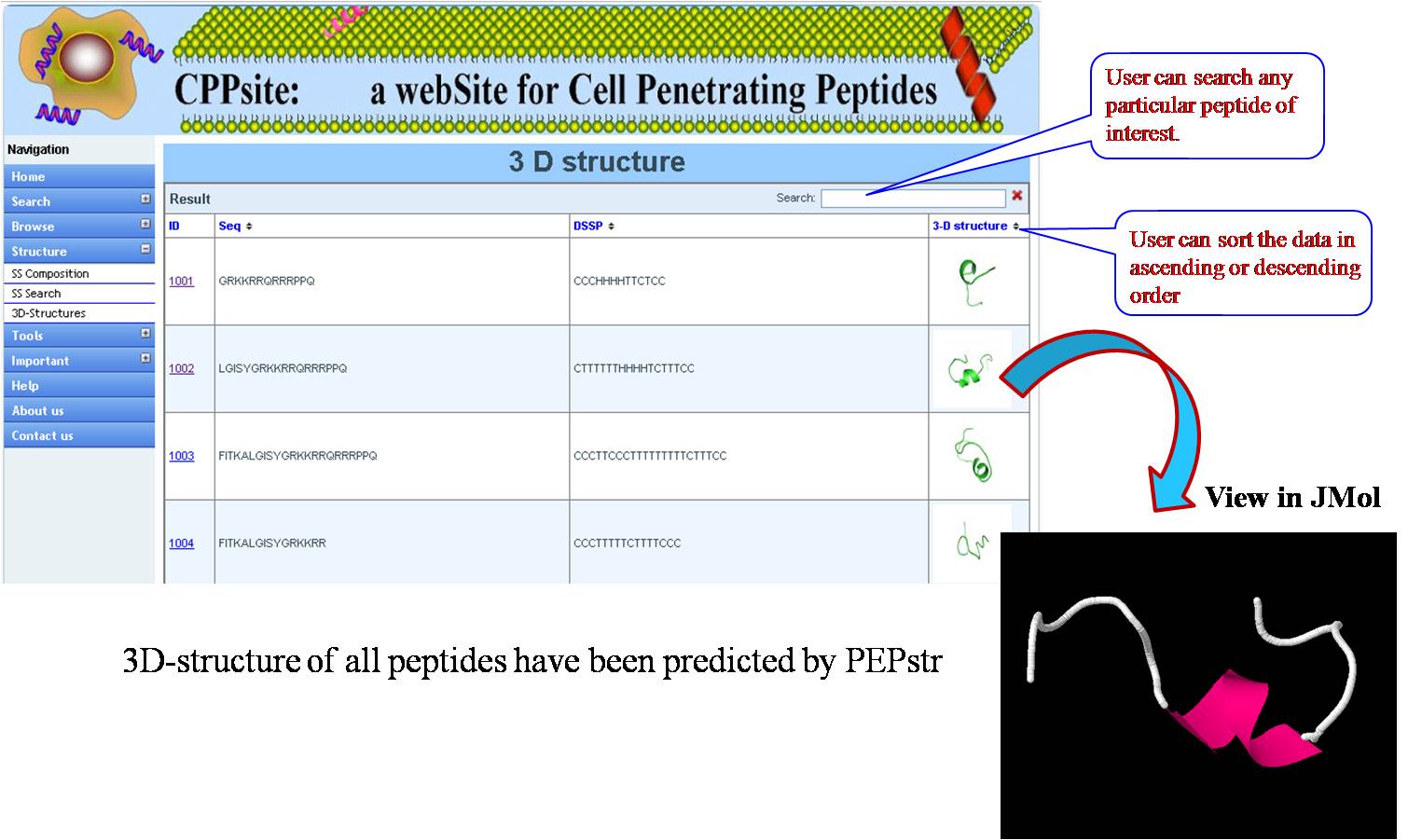
3D STRUCTURES 3D structures of all tumor homing peptides have been predicted using PEPstr. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
TOOLS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
BLAST It helps the user to run BLAST against Tumor homing peptide database. This is similarity based search of any query sequence with those present in tumor homing peptide database. User can submit query sequence in single letter code in search field, it will display all tumor homing peptides similar to query sequence. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
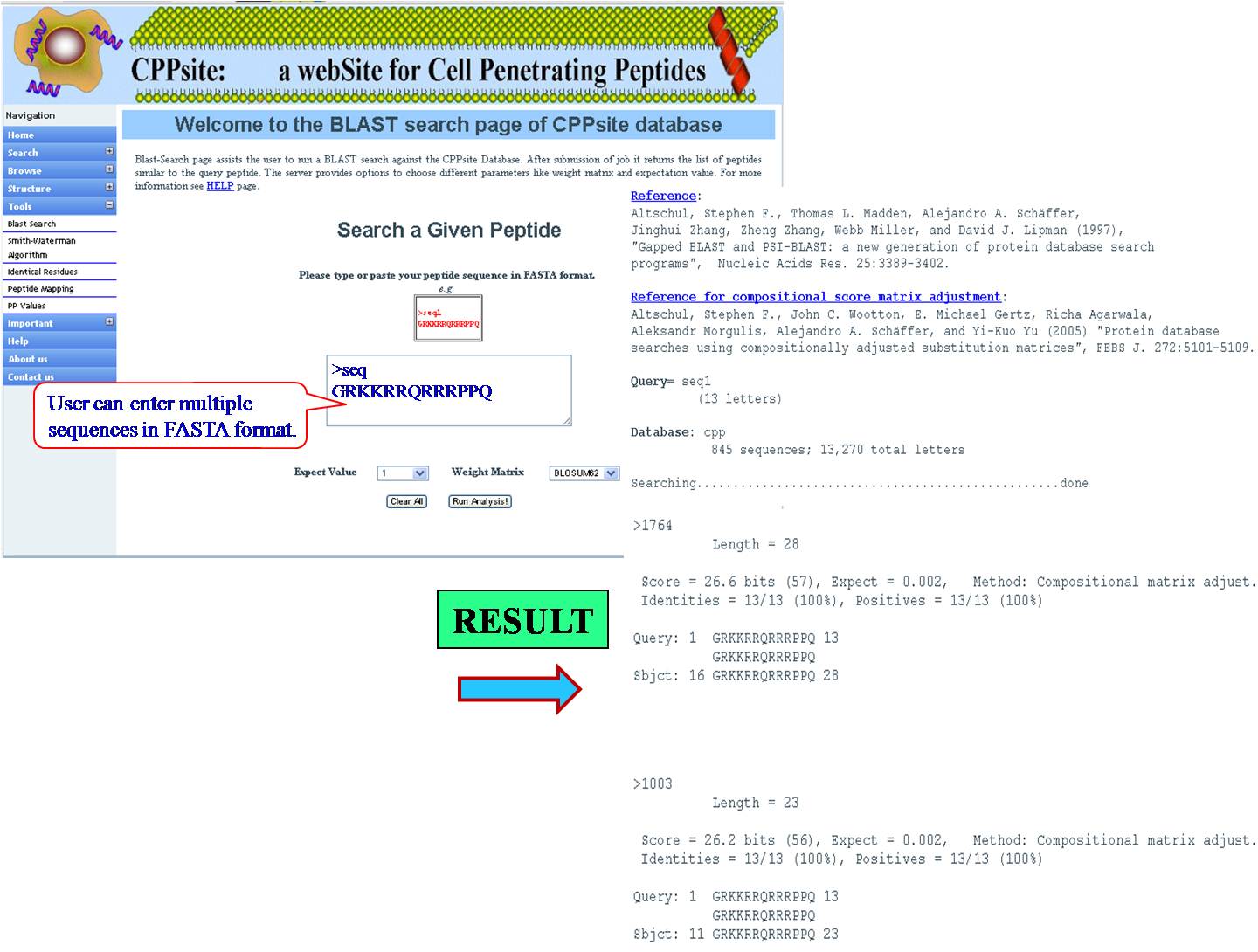 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
SMITH WATERMAN This tool can be used for determining similar regions between peptide sequences i.e. between query peptide and tumor homing peptides in the database, it compares the segments of possible lengths and optimizes the similarity measures giving exact result. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
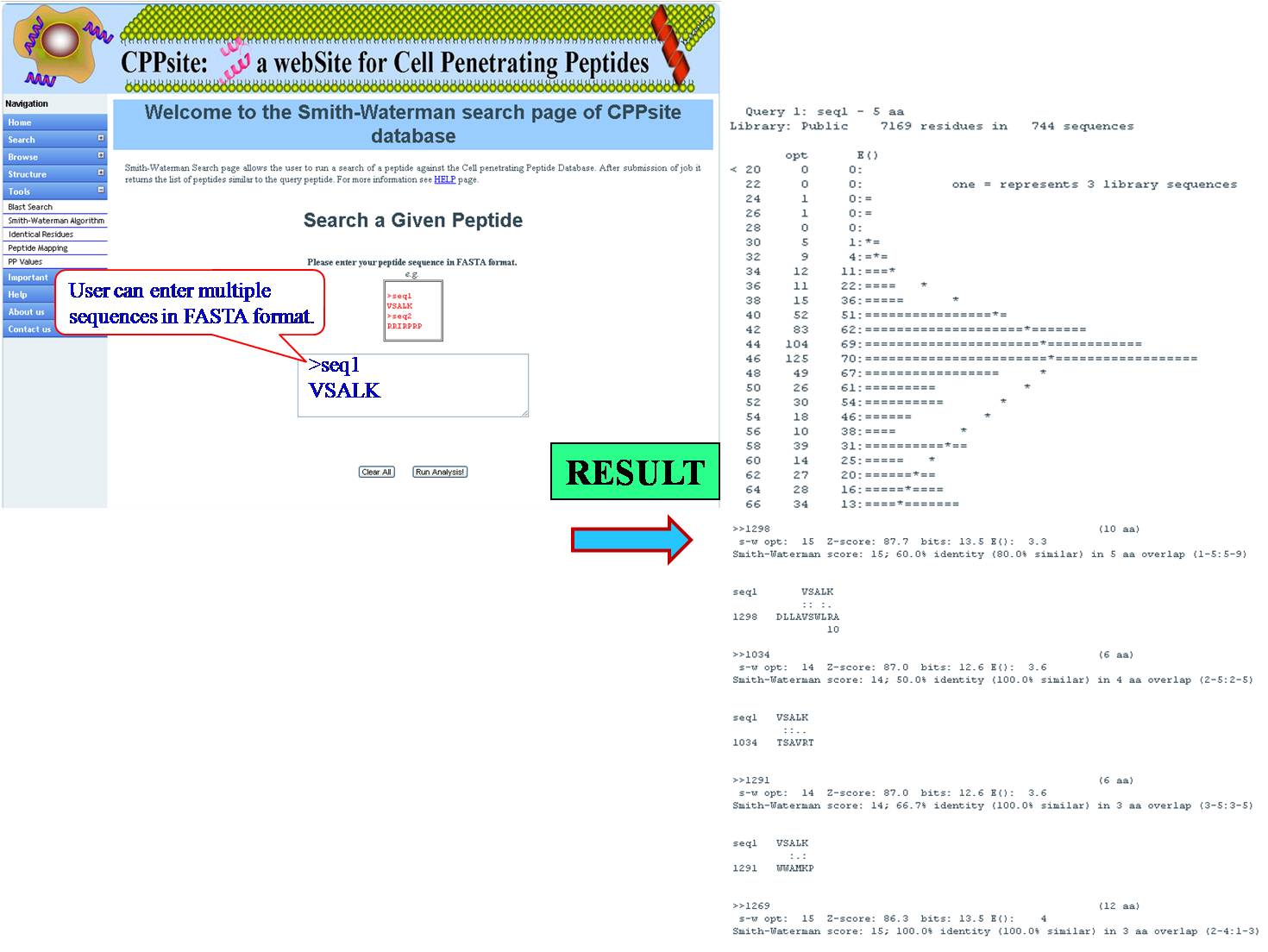 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
IDENTICAL RESIDUES Here the query sequence will be mapped stepwise against all the peptides of Tumor Homing Peptide database and number of exact matched residues of each pair will give Identical Residues value as final output for that pair. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
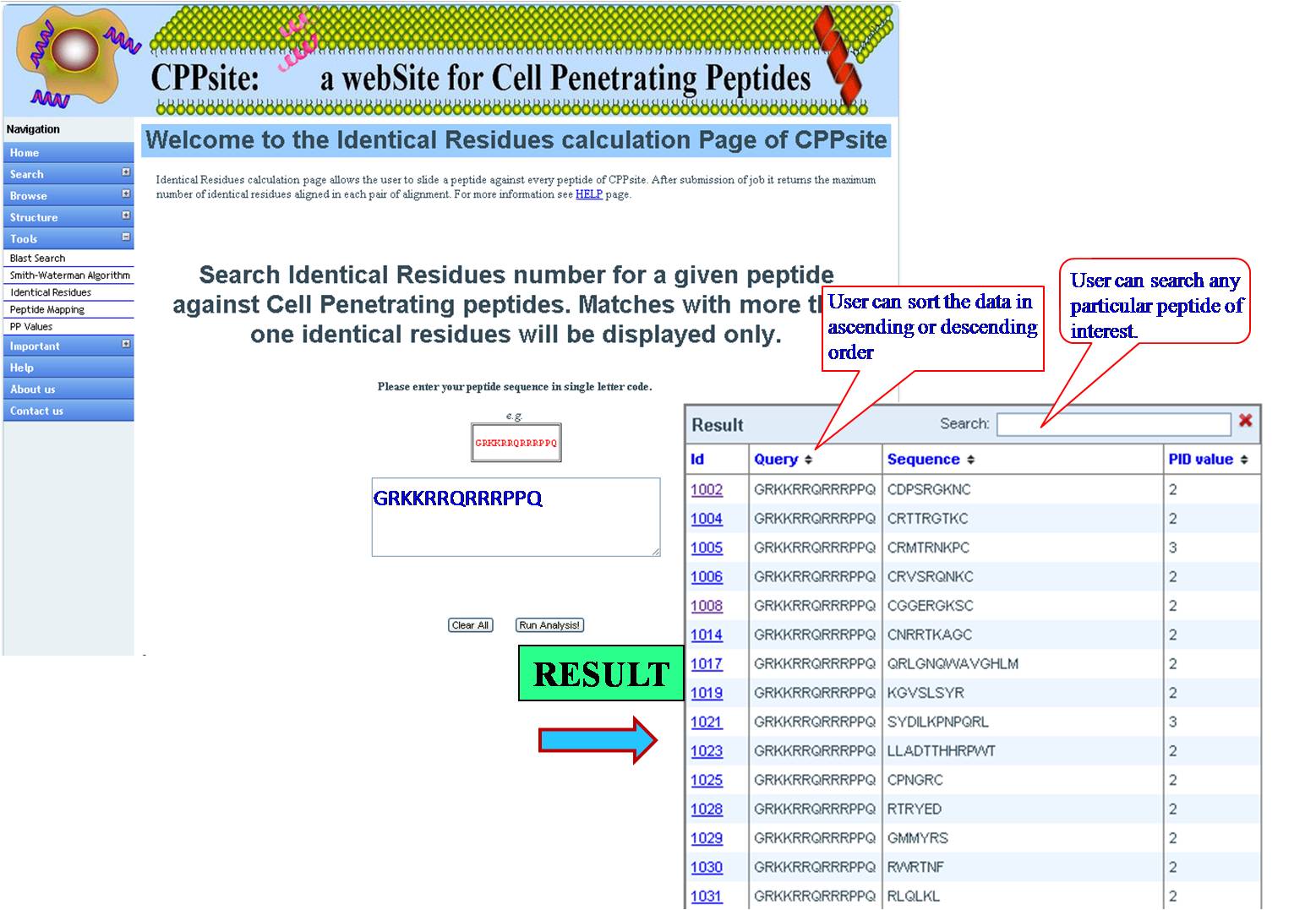 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
PEPTIDE MAPPING Peptide mapping is to find out if the query sequence or its motif is present in tumor homing peptide database. It is done in two ways-- SUBSEARCH: Maps the small peptide query against the tumor homing peptide database. SUPERSEARCH: Maps the tumor homing peptide database against large peptide/protein query. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
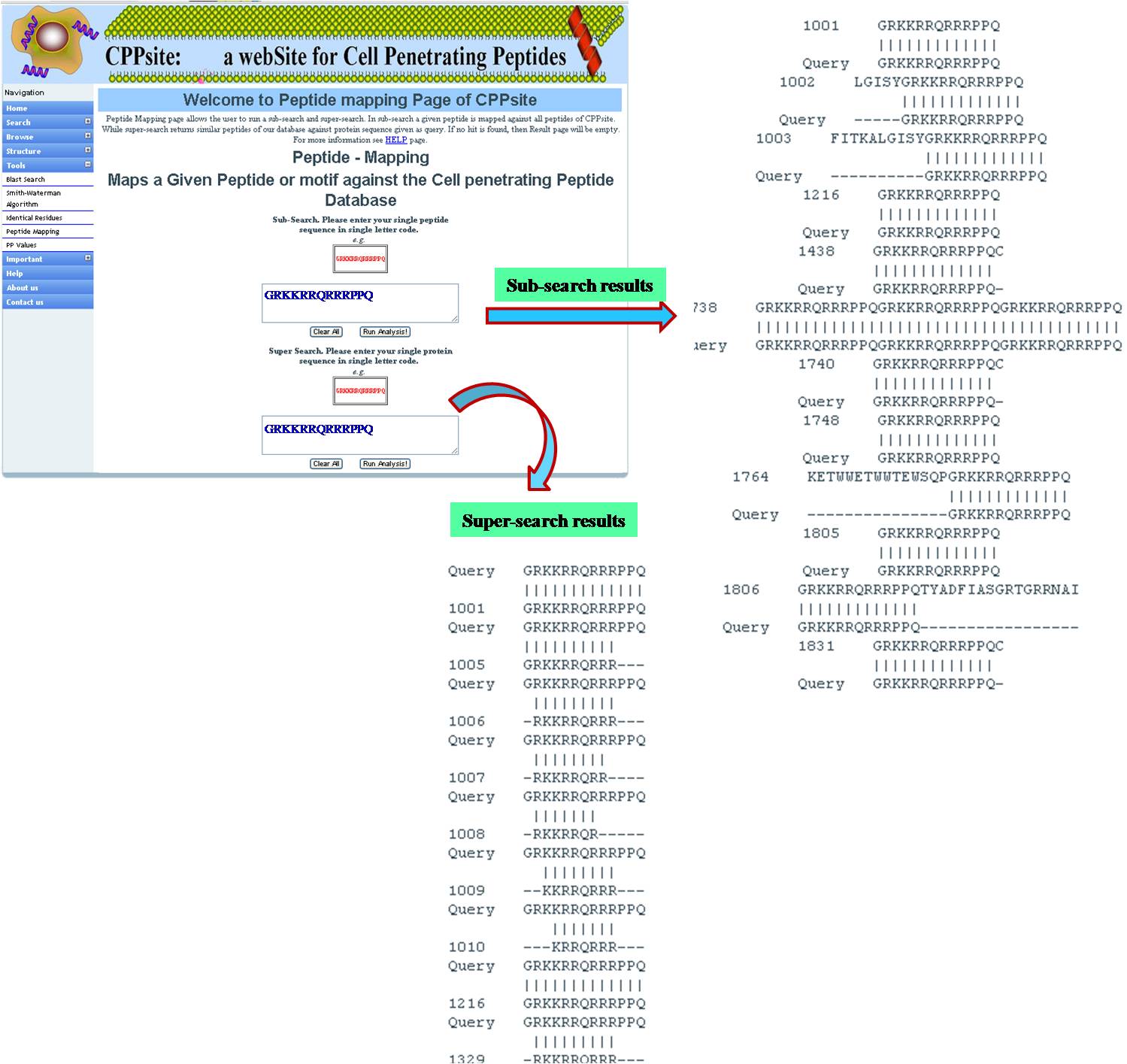 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
PHYSICAL PROPERTIES User gets physiochemical properties (hydrophobicity, hydrophilicity, Net charge, Iso electric point, Molecular weight) values for the query sequence. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
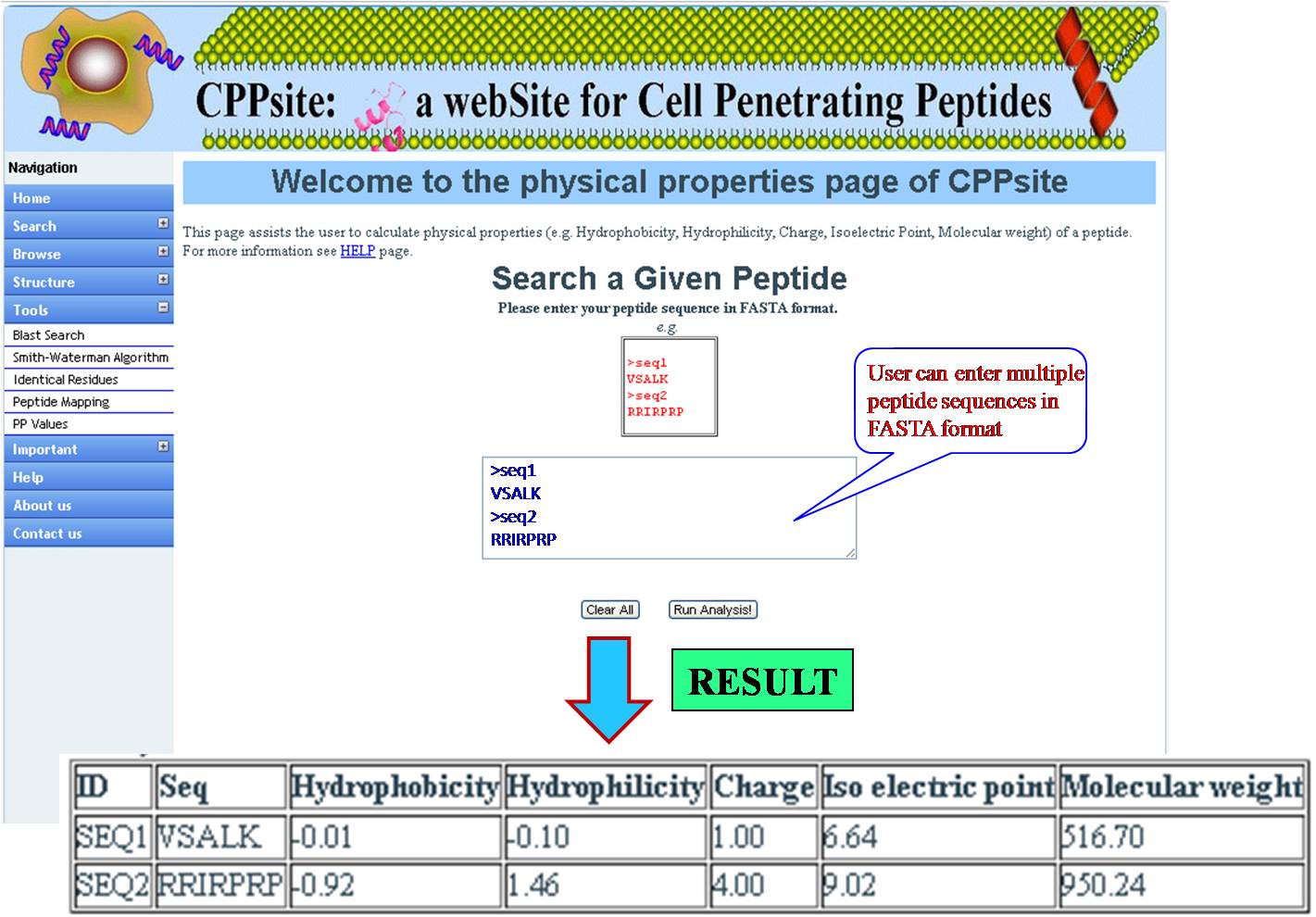 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
DESCRIPTION OF TABLE AND FIELDS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
| * In case of mutants (subcellular localization & uptake efficiency has not been determined), subcellular localization & uptake efficiency has been assumed similar to wild type (word 'probably' has been used). * Uptake efficiency has been provided in three categories: low (below 30% relative to control), medium (between 31-75% relative to control) and high (more than 75% relative to control). |
Frequently Asked Questions (FAQs)
|
Q1. What is CPPsite? Ans . CPPsite is a manually curated database of experimentally validated CPPs. Q2. Why CPPsite is created? Ans.CPP is one of the most promising tools for intracellular delivery of biological molecules and thus, has huge therapeutic potential. To the best of our knowledge, in spite of wealth of information on CPPs, no such database exists at present. Therefore, to help researchers who are working in the field of peptide based drug discovery, we have developed a comprehensive database which provides all the basic information about CPPs. Q3.What is unique about CPPsite? Ans. CPPsite is a first and unique database of its kind, which has many online tools. One of the powerful tools is peptide mapping and similarity search, which allows user to search similar peptides/motifs in a query sequence. In addition, CPPsite also provides 3D structures of CPPs. Q4. Does this database represent all experimentally validated CPPs? Ans. Ans.No. This database is the result of first round curation and we shall update CPPsite on a rolling basis, regularly adding CPPs from literature. Q5. Why search CPPsite? Ans. CPPsite provides all experimental information of used CPPs such as uptake efficiency, uptake mechanism, hydrophobicity, sub-cellular localization, N- and C- terminal modifications, cell lines used etc. This information may be very useful for researchers working in these areas. Information about the structural content of CPPs may be useful for designing of novel CPPs. Apart from this; user can search CPPs having high uptake efficiency or can use various tools available to calculate AA frequency, AA composition and hydrophobicity of a query sequence. Q6. How do I process a text search with CPPsite database? Ans: You can process any name, or ID search through the CPPsite homepage or you may go to complex search for specific queries. Q7. To whom can I report a discrepancy? Ans. Please refer to the "Contact Us" page. |



