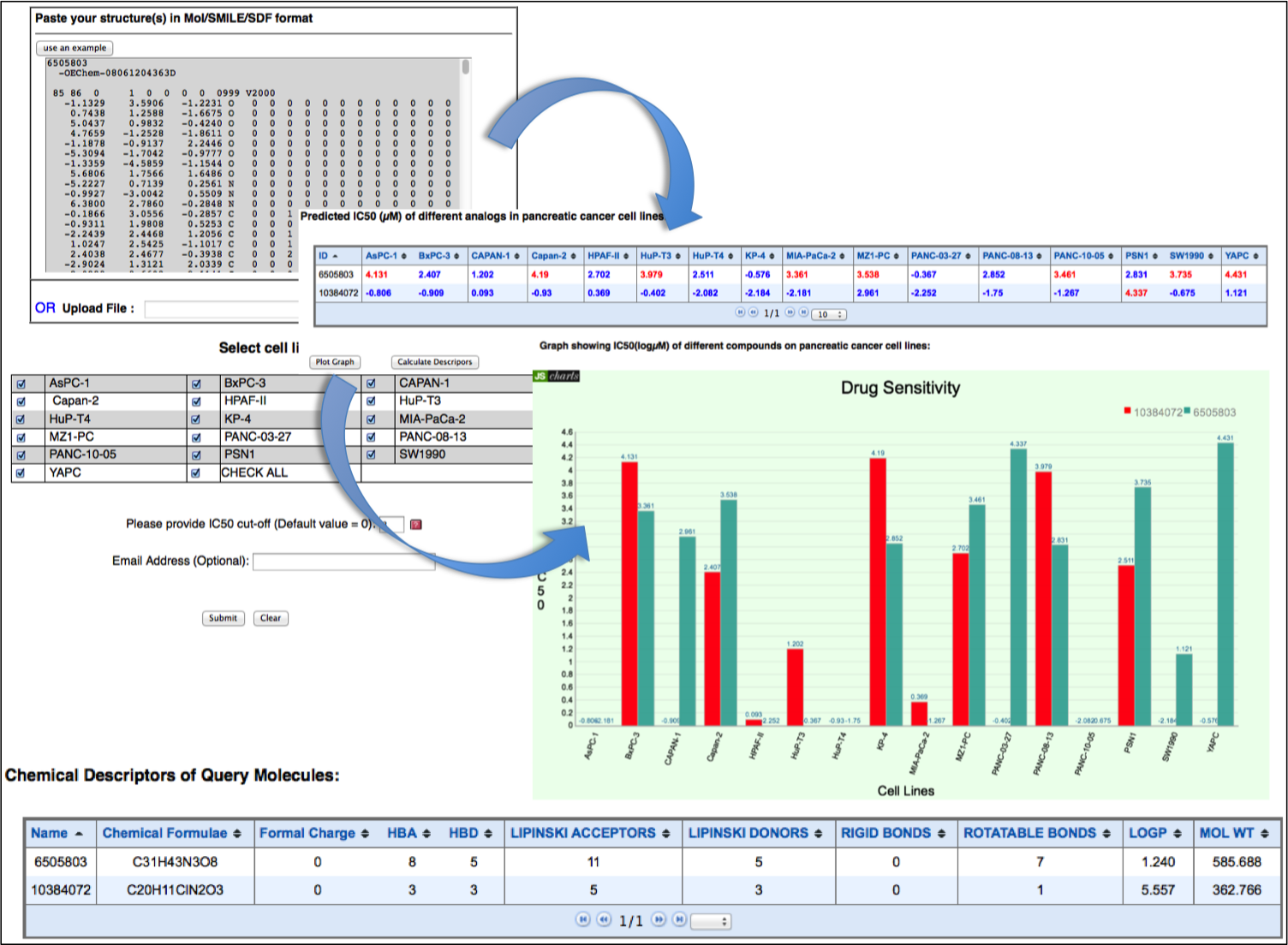
This tool allow user to draw chemical structure of their molecule by using JME editor. At one time user can predict drug sensitivity on maximum of 16 pancreatic cancer cell lines. It is very difficult to define the cut-off IC50 value, which discriminate sensitive and resistance cancer cell lines. So, we provided an option of IC50 cut-off value, which will be define by the user on the basis of their experimental criteria. On submission, DiPCell returns with logIC50 values on pancreatic cancer cell line chose by the user and with an option to calculate chemical descriptors of the query molecule. |
In medicinal chemistry, it is always often that analog of certain molecule is more active than parent one. So, it is a common practice to identify a better molecule of a certain existing drug by structural activity relationship (SAR). In DiPCell, we have incorporated the same kind of a module, where user can design analogs and simultaneously predict their drug sensitivity on pancreatic cancer cell lines. Further, user can choose the best analog and generate their analogs for further refinements. User has to provide scaffold structures, building blocks and linkers as input for this module. |