Algorithm for DMKPred
SVM
Evaluation Modules
The performance modules constructed in this study were evaluated using a leave one out cross-validation (LOOCV) technique. In the leave one cross-validation, the relevant dataset was randomly divided into two sets, testing and training set. The training and testing was carried out N times, each time using one distinct set for testing and the remaining sets for training, where N is the total number of molecules.We computed the correlation between predicted and actual binding of chemical kinase inhibitors.
Pearson correlation coefficient between the actual and the predicted binding of chemical kinase inhitors with protein kinases was calculated to assess the performance of our real binding prediction modules. The Pearson correlation coefficient (r) between the actual and the predicted efficacies has been calculated using following equation. The performance of the methods was computed using the following formulas
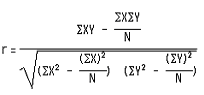
Where r is Pearson's correlation coefficients, X is the actual binding, Y is SVM predicted binding and N is total number of examples.
The performance modules constructed in this study were evaluated using a leave one out cross-validation (LOOCV) technique. In the leave one cross-validation, the relevant dataset was randomly divided into two sets, testing and training set. The training and testing was carried out N times, each time using one distinct set for testing and the remaining sets for training, where N is the total number of molecules.We computed the correlation between predicted and actual binding of chemical kinase inhibitors.
Pearson correlation coefficient between the actual and the predicted binding of chemical kinase inhitors with protein kinases was calculated to assess the performance of our real binding prediction modules. The Pearson correlation coefficient (r) between the actual and the predicted efficacies has been calculated using following equation. The performance of the methods was computed using the following formulas
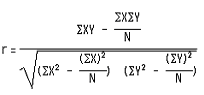
Where r is Pearson's correlation coefficients, X is the actual binding, Y is SVM predicted binding and N is total number of examples.
Bioinformatics Center, Institute Of Microbial Technology
Chandigarh,India