 ParaPep -A Database of Anti-parasitic peptides
ParaPep -A Database of Anti-parasitic peptides
Acknowledgments |
| We are grateful to researchers for their direct and indirect help in the development of this database. In this database we used information from various resources/databases as well as we used number of software/web services for developing this database. We are grateful to the developers of these databases/software/resources that include following resources. |
| Link | Description |
|---|---|
OSRA | Optical Structure Recognition Application. |
BLAST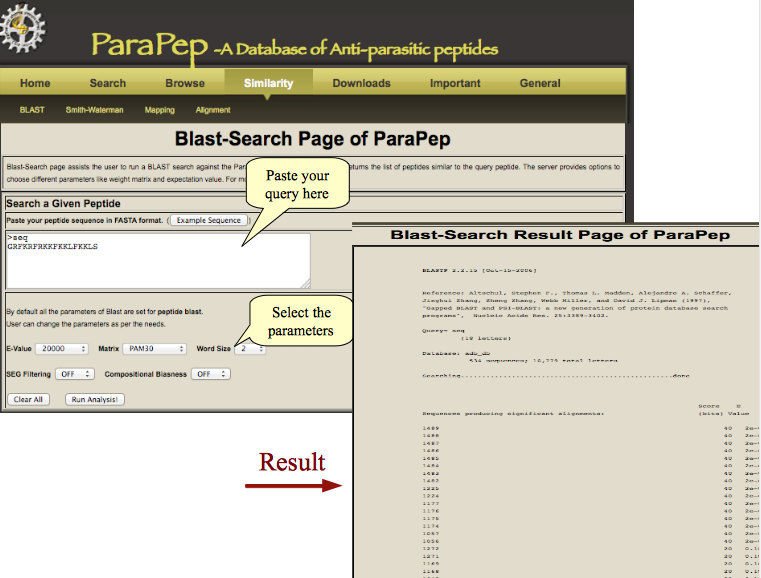 | Algorithm for pairwise similarity search. |
CLUSTALW2 | Algorithm for multiple sequence alignment. |
DSSP | Database for secondary structure assignments. |
IEDB | Database of antibody and T cell epitopes for humans, non-human primates, rodents, and other animal species. |
JALVIEW | Program for editing, visualization and analysis of multiple sequence alignment. |
Jmol | An open-source Java viewer for chemical structures in 3D. |
MUSTANG | Program for multiple structural alignment of proteins. |
PyMOL | PyMOL is a user-sponsored molecular visualization system on an open-source foundation. |
UCSF chimera | UCSF Chimera is a highly extensible program for interactive visualization and analysis of molecular structures and related data. |
ChemDraw | ChemDraw is the drawing tool of choice for chemists to create publication-ready, scientifically intelligent drawings for use in ELNs, databases and publications and for querying chemical databases |