|
|||
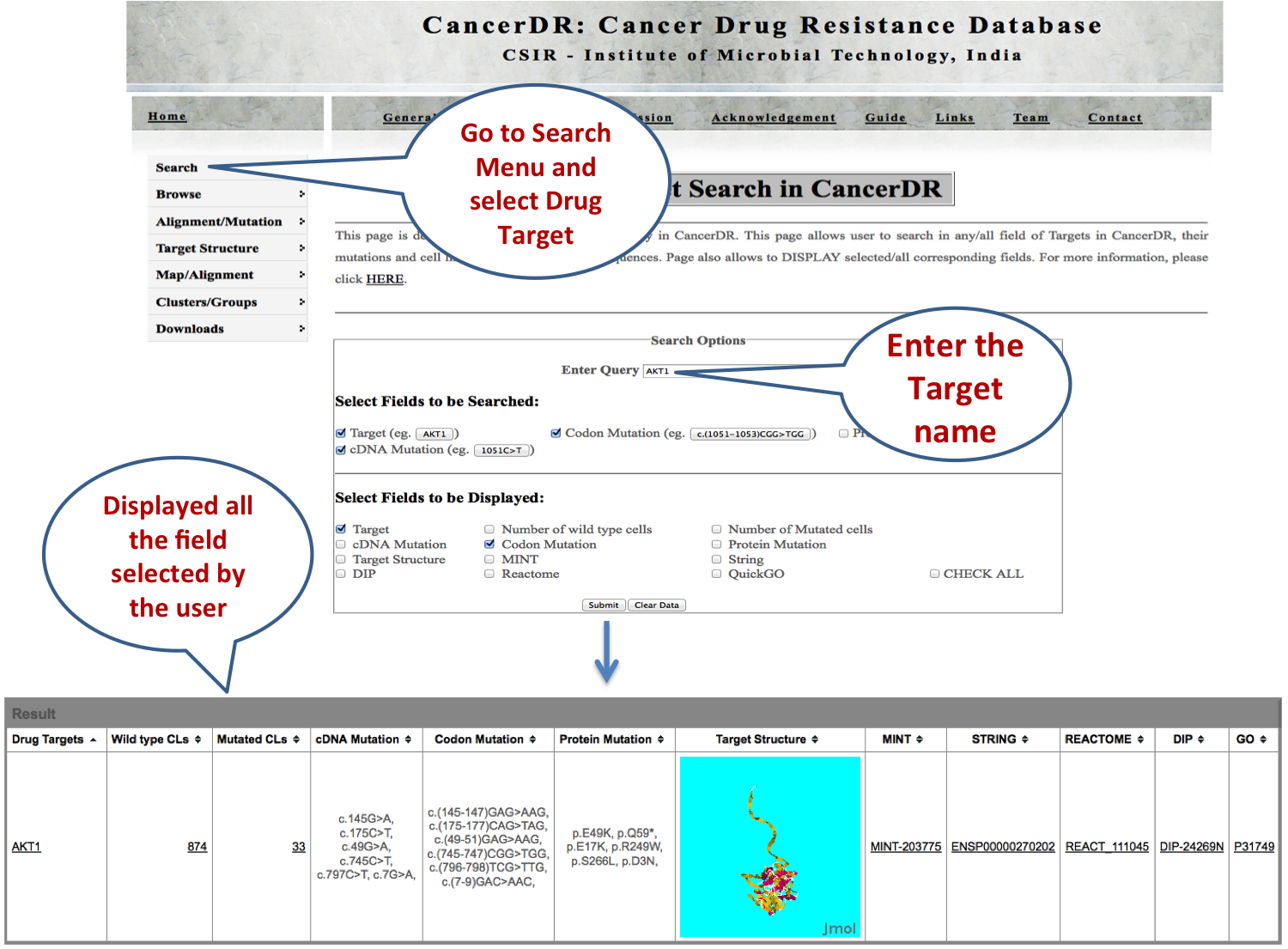
SEARCH |
DRUG TARGETS Target search option allows the user to obtain detailed information about a drug target including the number of cell lines having mutated or normal drug target, protein change, cDNA change, codon change and the predicted structure of the target. The links allow user to get relevant information of drug target like protein interactions, pathway interactions and gene ontologies. |
 |
CELL LINES Cell line search can be done to get the tissue type to which the cell line belongs. The other column tells about the number of drugs those have been tried against that cell line. |
 |
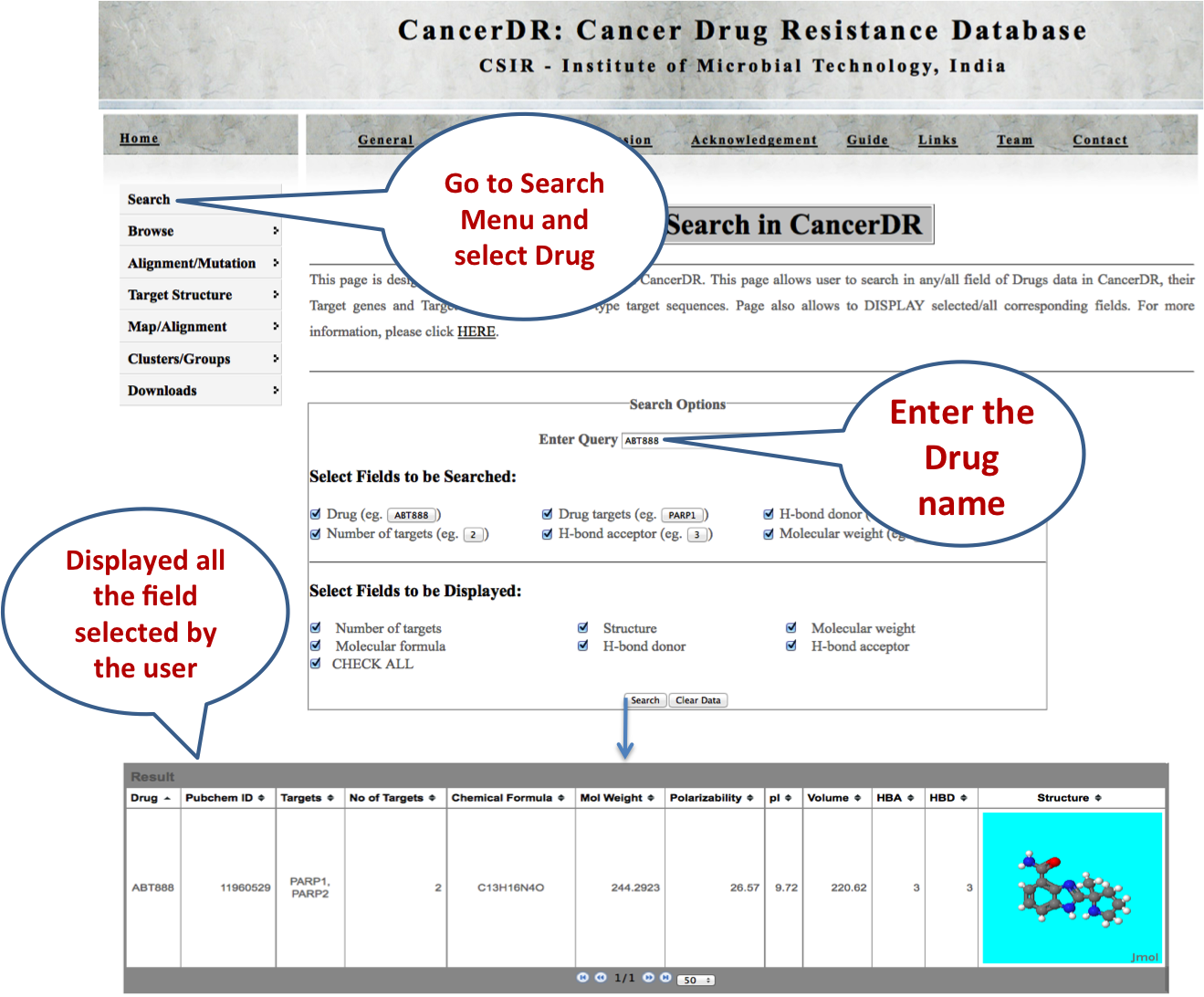
DRUGS Drug search enables the user to find the structure of the drug molecule along with other molecular details. User can also search the targets of drugs and number of targets. |
 |
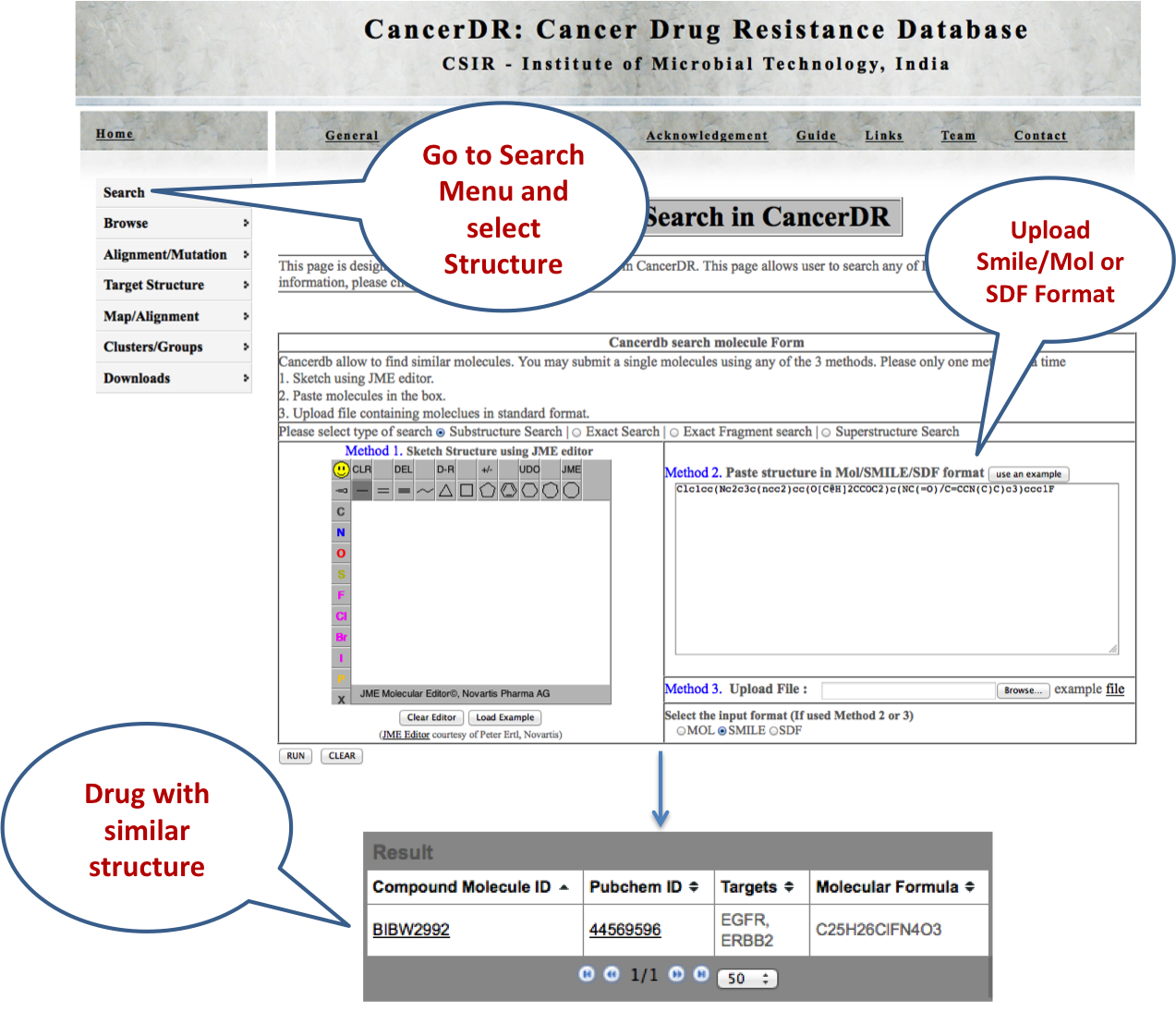
STRUCTURE Here user can search the CancerDR drugs on the basis of similarity. They can use SDF, MOL or SMILE format for their search. In addition to this, user can also draw the molecules. |
 |
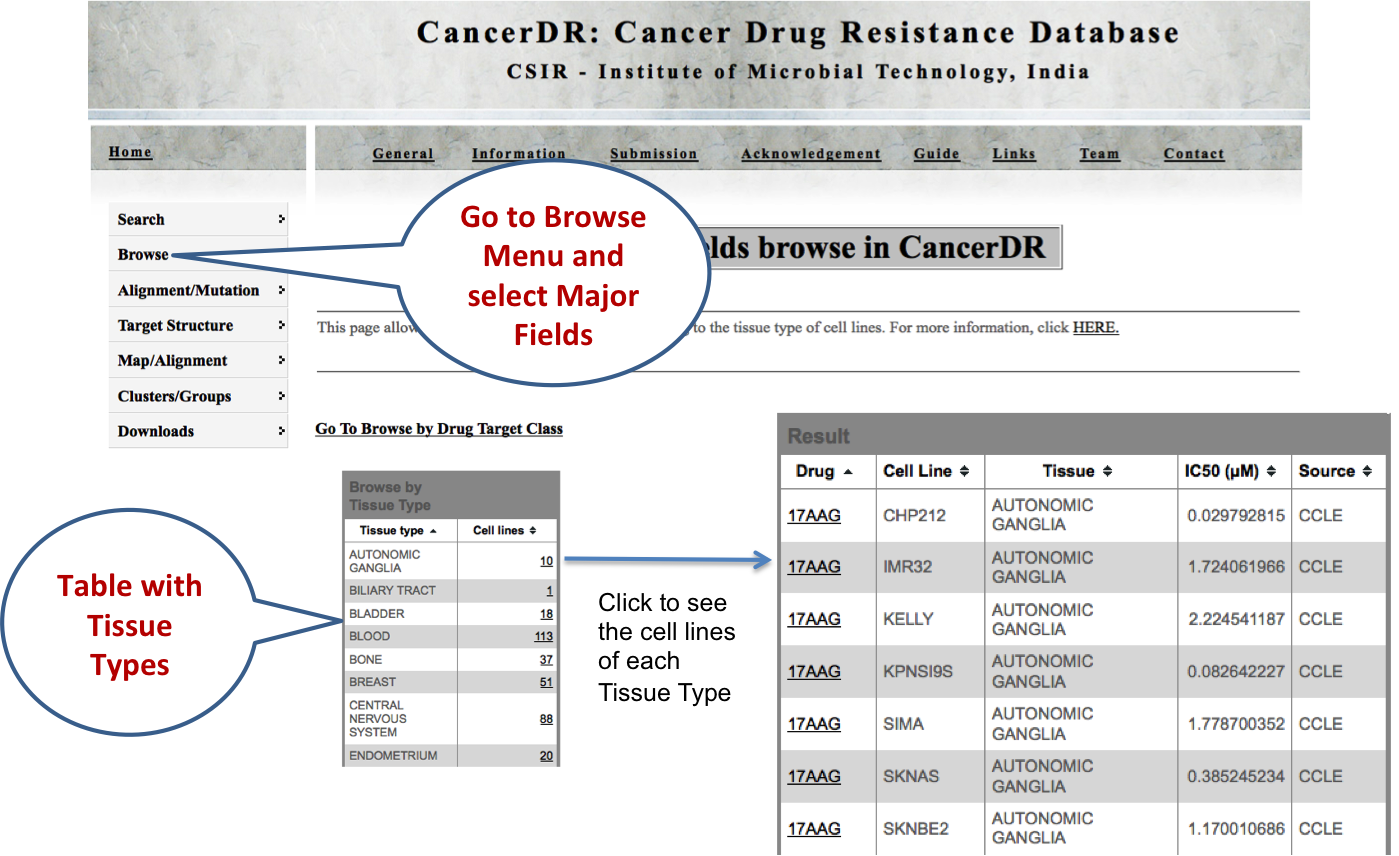
BROWSE |
MAJOR FIELDS Here user can browse the CancerDR data on the basis of two major categories. First, tissue type of cell lines used in pharmacological drug profiling. Second, molecular therapeutic targets of the anticancer drugs. |
 |
 |
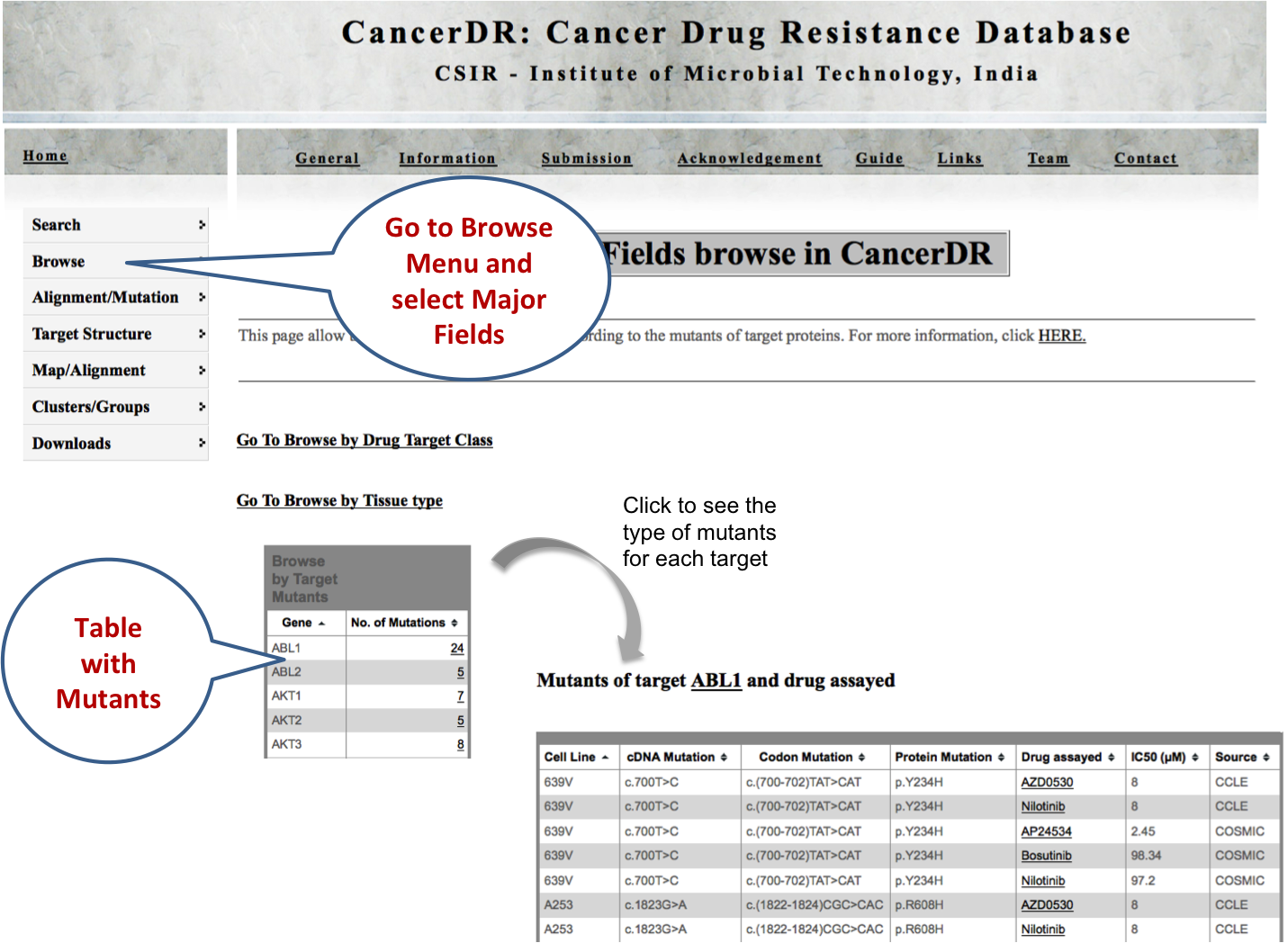 |
DRUG TARGETS Drug target browsing facilitates the user to get the comprehensive information of drug targets such as links for mutations, protein-protein interactions, pathway interactions, gene ontologies, genome browser, phylogenetic tree, chromosomal position and number of cell lines with mutated drug targets. |
 |
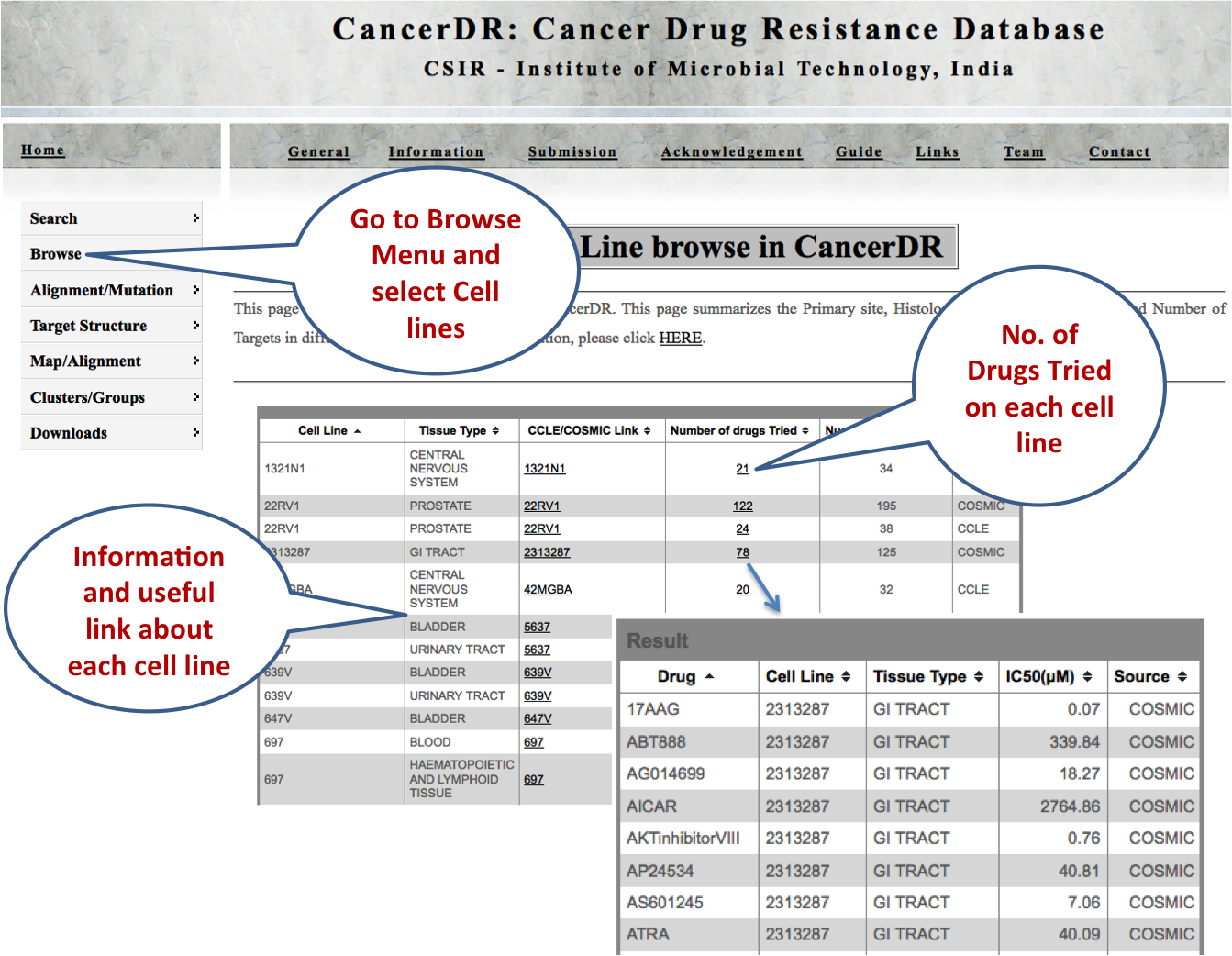
CELL LINES This browsing option enables the user to look at the number of drugs and their targets along with the information regarding the histology of the cell lines. |
 |
DRUGS With this option, the user can look at different descriptors of drugs like pI, Volume, Hydrogen Bond Acceptor, Hydrogen Bond Donor, Structure link, molecular target, along with other chemical properties and structures of drugs. |
 |
ALIGNMENT/MUTATION |
TOTAL ALIGN Here user can do the multiple sequence alignment of drug targets. This is divided into two parts first, alignment of mutated sequences with wild type sequence of particular target. Second, alignment of natural variant sequences of targets available in Uniprot. Jalview is also integrated in this module to show the alignment in a better way to understand the sequence variation. |
 |
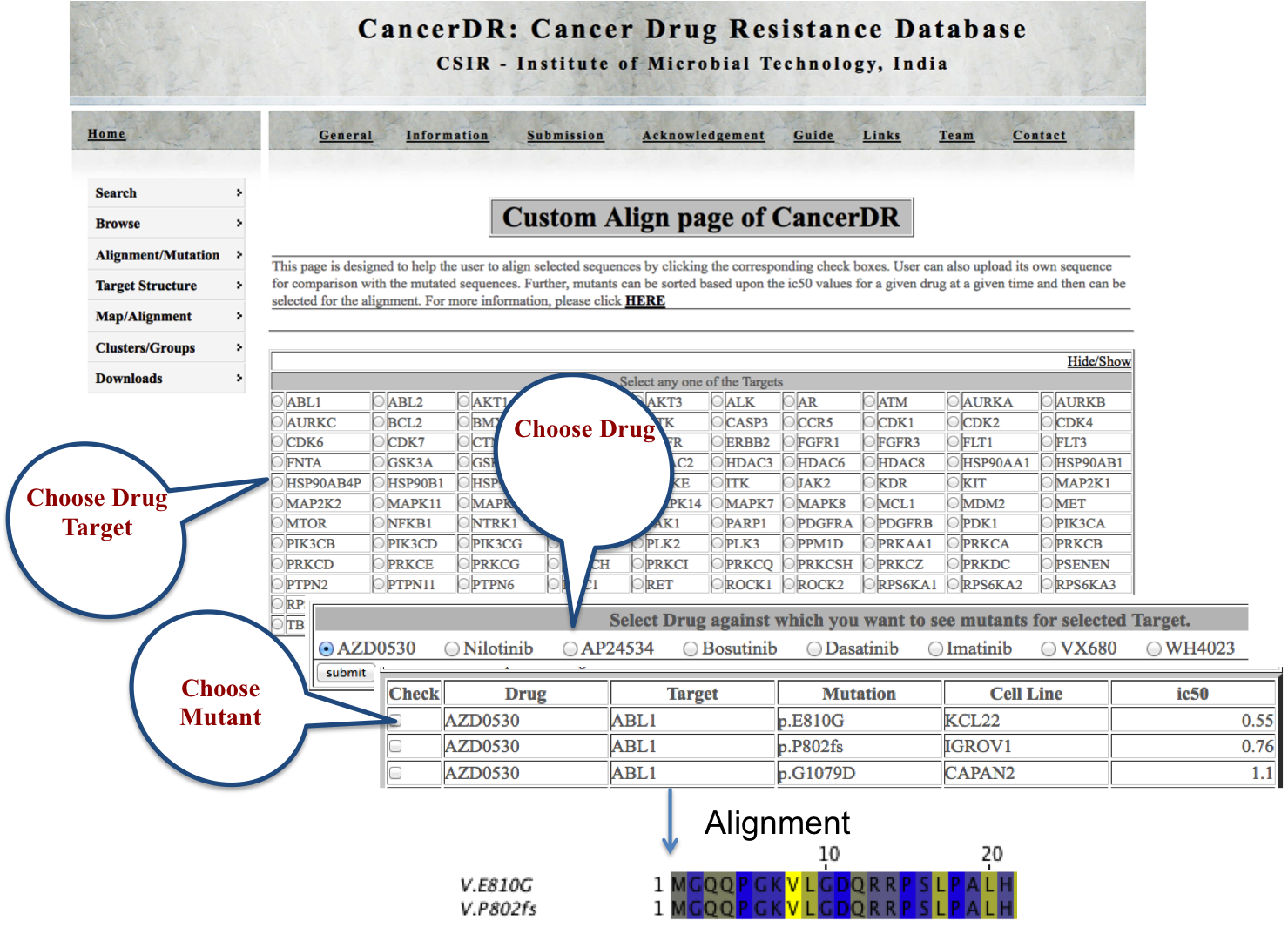
CUSTOM ALIGN Custom align module gives an advantage to the user to choose the mutated sequences of a particular target for multiple sequence alignment. Here user can correlate the mutated sequences with the IC50 values. |
 |
MUTANTS Here user can find out the how many mutations are present in particular drug targets. Mutations are available at three levels viz. cDNA, codon and protein. |
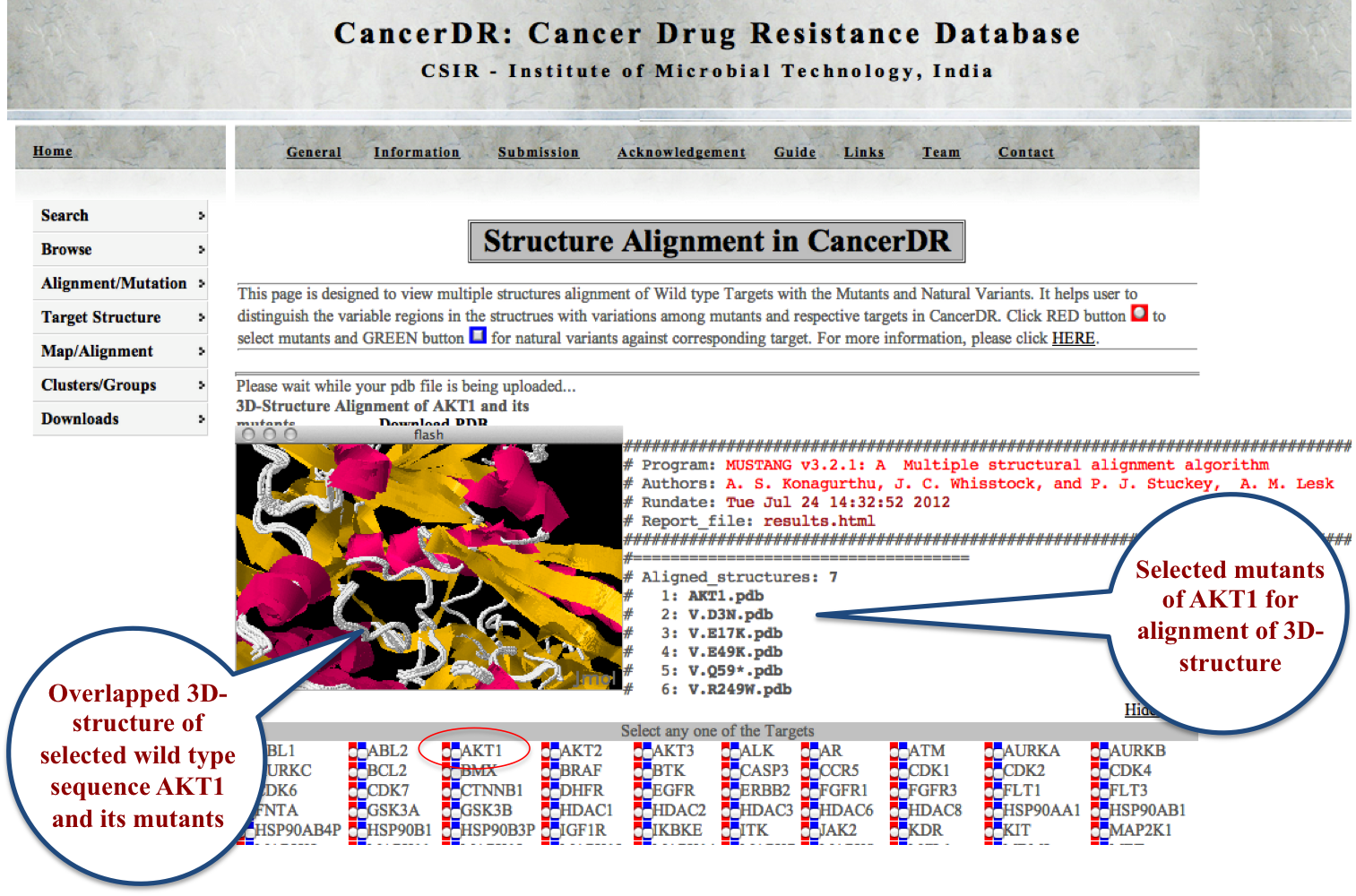
STRUCTURE ALIGNMENT This powerful module enables the user to align the tertiary structures of two or more mutants of a particular drug target to see the structural changes in respective mutants. Variants of drug targets can also be aligned here. Sequence alignment on the basis of structural alignment can also be generated over here. |
 |
TARGET STRUCTURE |
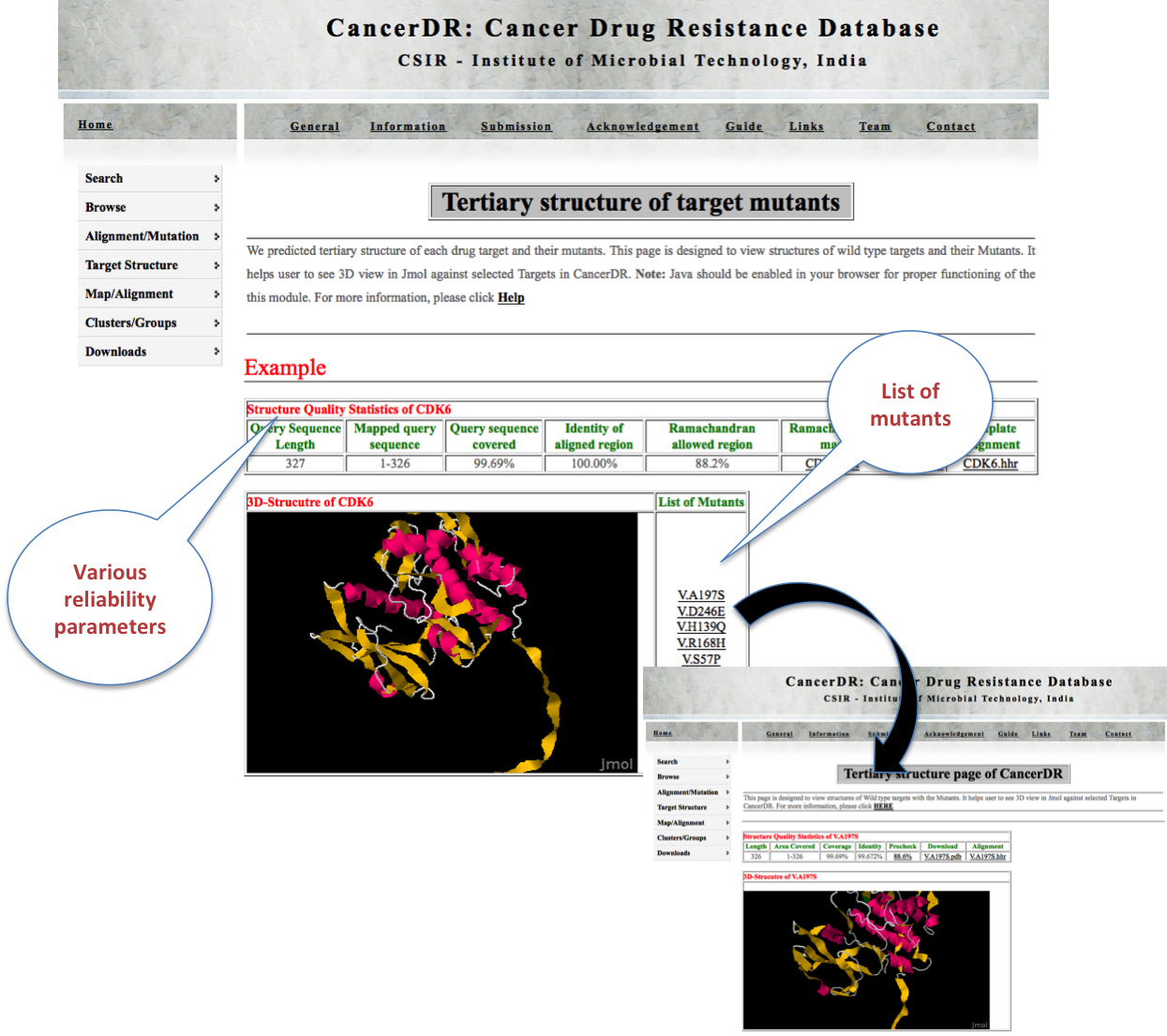
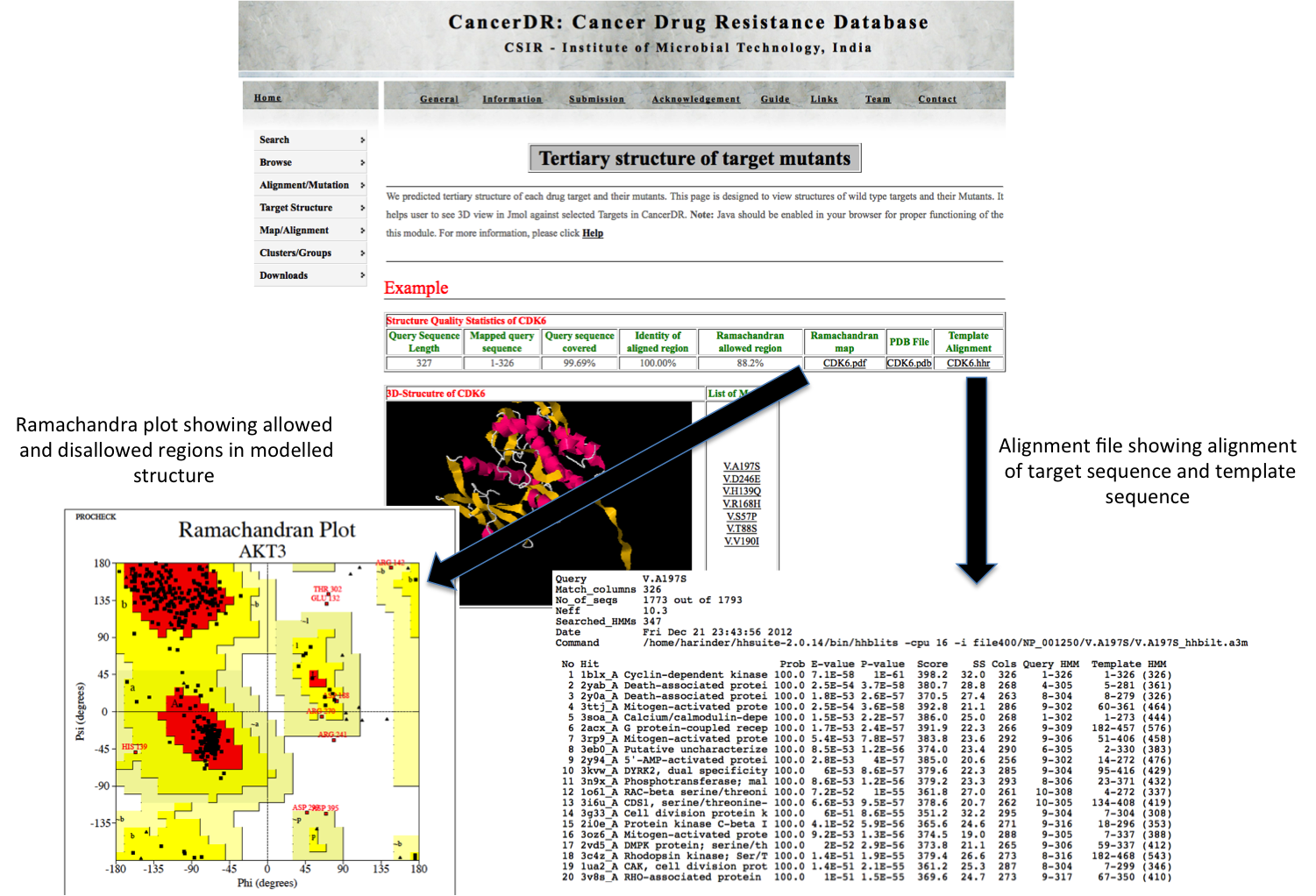
TERTIARY Here user can view the tertiary structure of drug targets and their mutants in Jmol. In each mutant, mutated position is depicted in green color to locate the mutation region. PDB file of each structure can also be downloaded from here. |
 |
 |
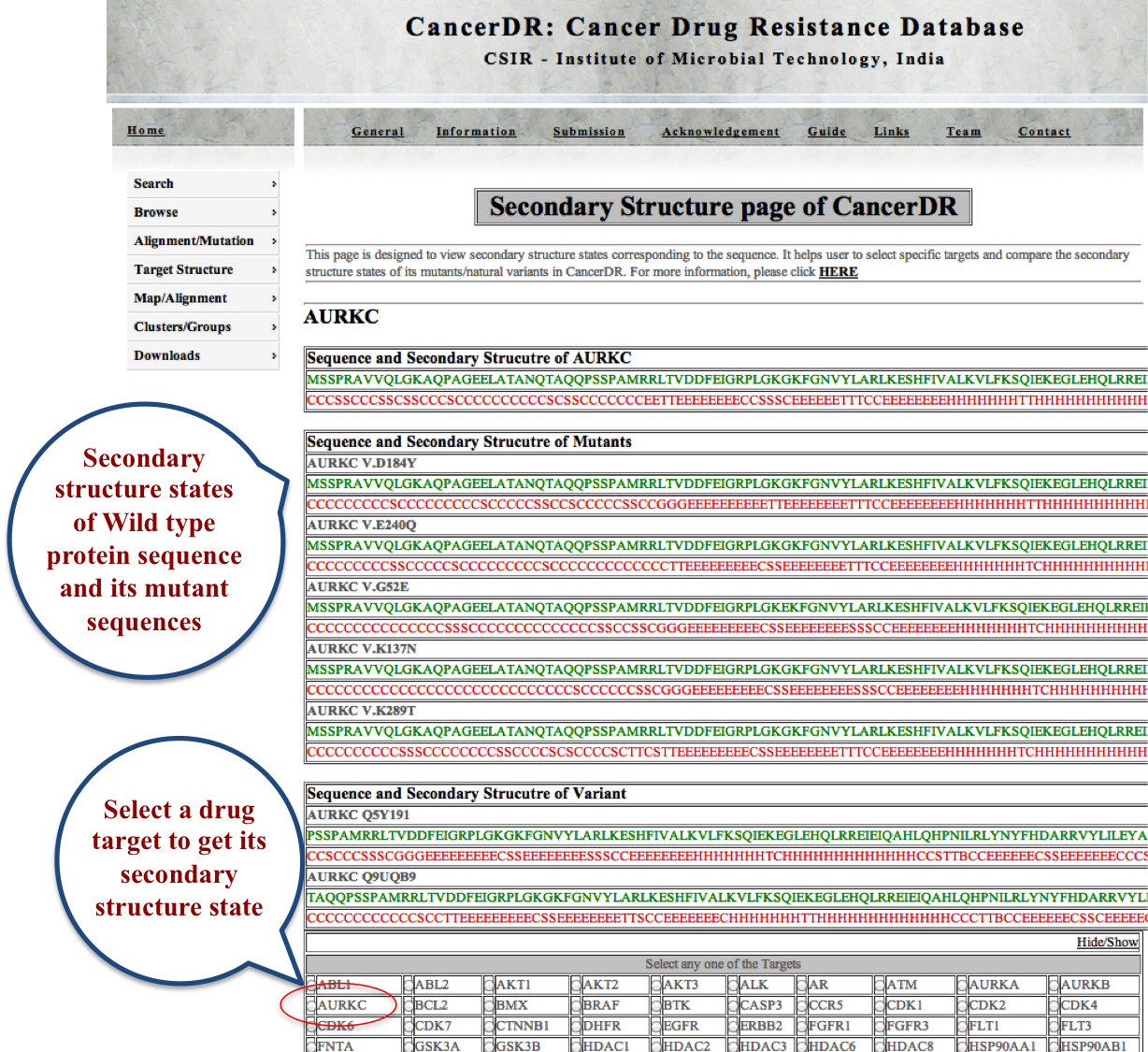
SECONDARY It is very important to know that which residue is present in which secondary state e.g. helix, beta sheet or turn. To keep this in mind, we obtained the secondary state of all the drug targets, mutants and natural variants by using DSSP. |
 |
COMPARE Compare module is built to compare the structure of two mutants of a particular drug target. Users can also compare their own structures with the any structure present in CancerDR. |
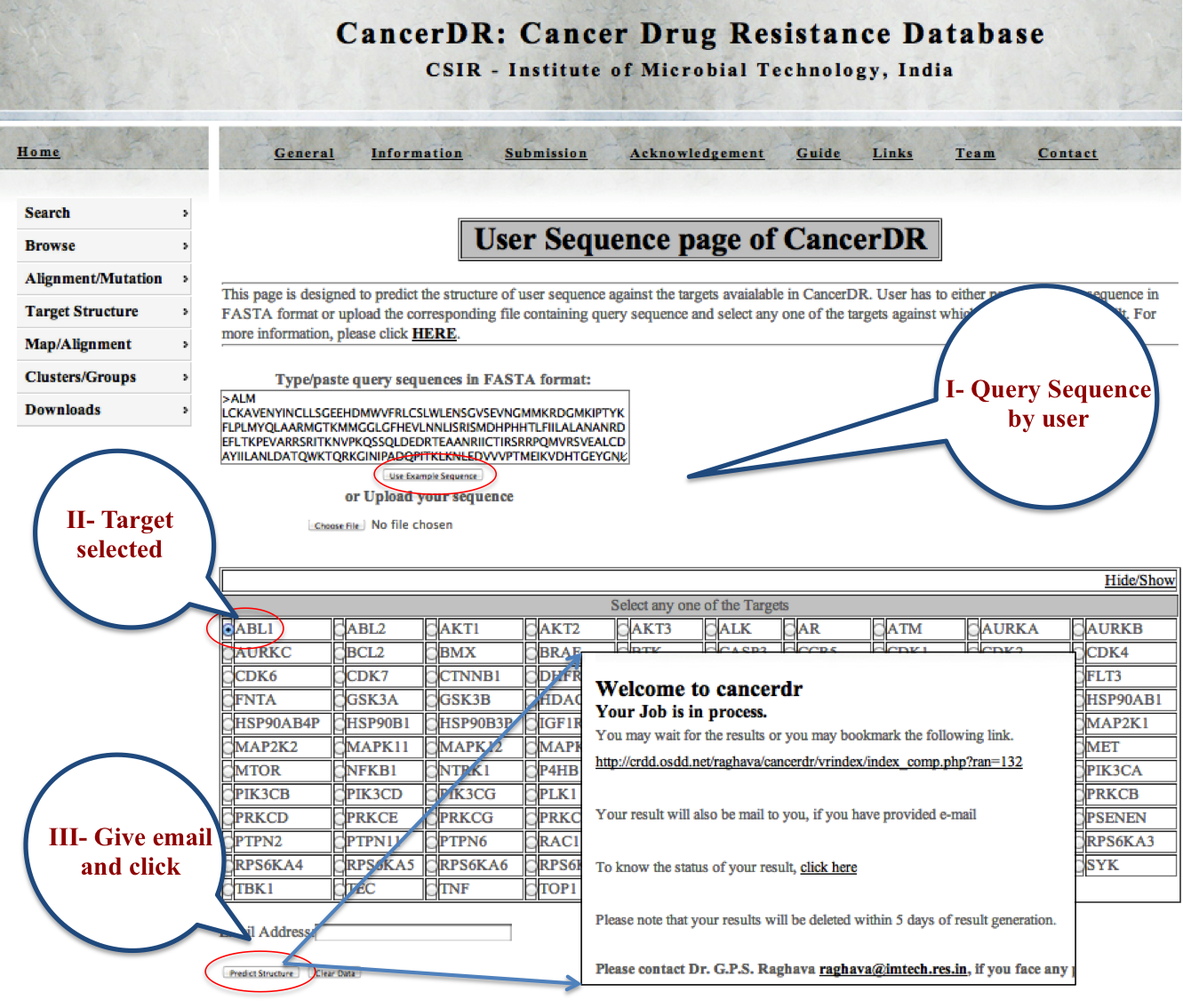
USER SEQUENCE This module is designed to predict the structure of user sequence against the targets available in CancerDR. User has to either paste the query sequence in FASTA format or upload the corresponding file containing query sequence and select any one of the targets against which structure has to be built. |
 |
MAP/ALIGNMENT |
Short Reads This module will be used for the alignment of short reads, produced after the Next Generation Sequencing (NGS) of Transcriptome or Exome. Both types of sequencing reads i.e Paired end and single end sequencing reads can be submitted to this module. Sequencing reads will be aligned to the Cancer target genes and visualized for any variation. Alignment visualization will clearly indicate the mutations in the sequenced genes. User has to provide the files (.fastq format) generated after the sequencing (Please see the example files). This module use BWA to generate alignment files (i.e. aln.sam file) and Tablet for visualization of the alignment files. It is necessary to provide forward and reversed read files both at a time if a user wants to use paired end data for the alignment. A single file is enough in case of single end sequencing. Please have a look at the help pictures. |
| Help Pictures |
Contigs In this module user can submit contigs (Genomic or transcriptomic) for the alignment of genes (extracted from those contigs) with Cancer targets. This module works in two steps. In the first step, genes have been predicted from the contigs with Augustus software. After that the predicted genes (nucleotide and proteins sequences both) will be searched for similarity with Cancer targets by BLAST. At the result page, user can analyze the Predicted genes and their alignment with cancer targets. Please have a look at the help pictures. |
| Help Pictures |
Sequences |
| Help Pictures |
CLUSTERS/GROUPS |
TARGETS |
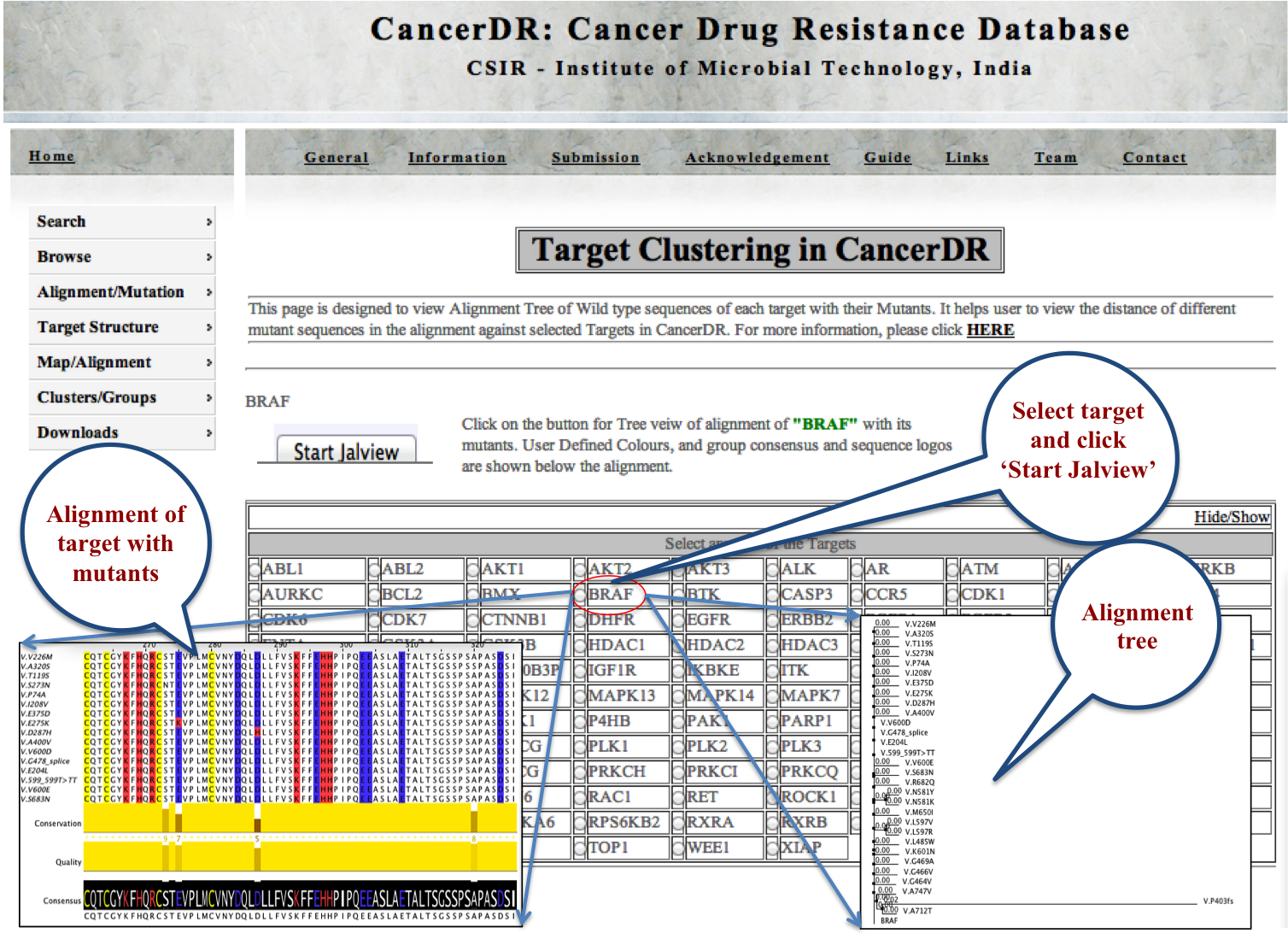 |
Cell Lines |
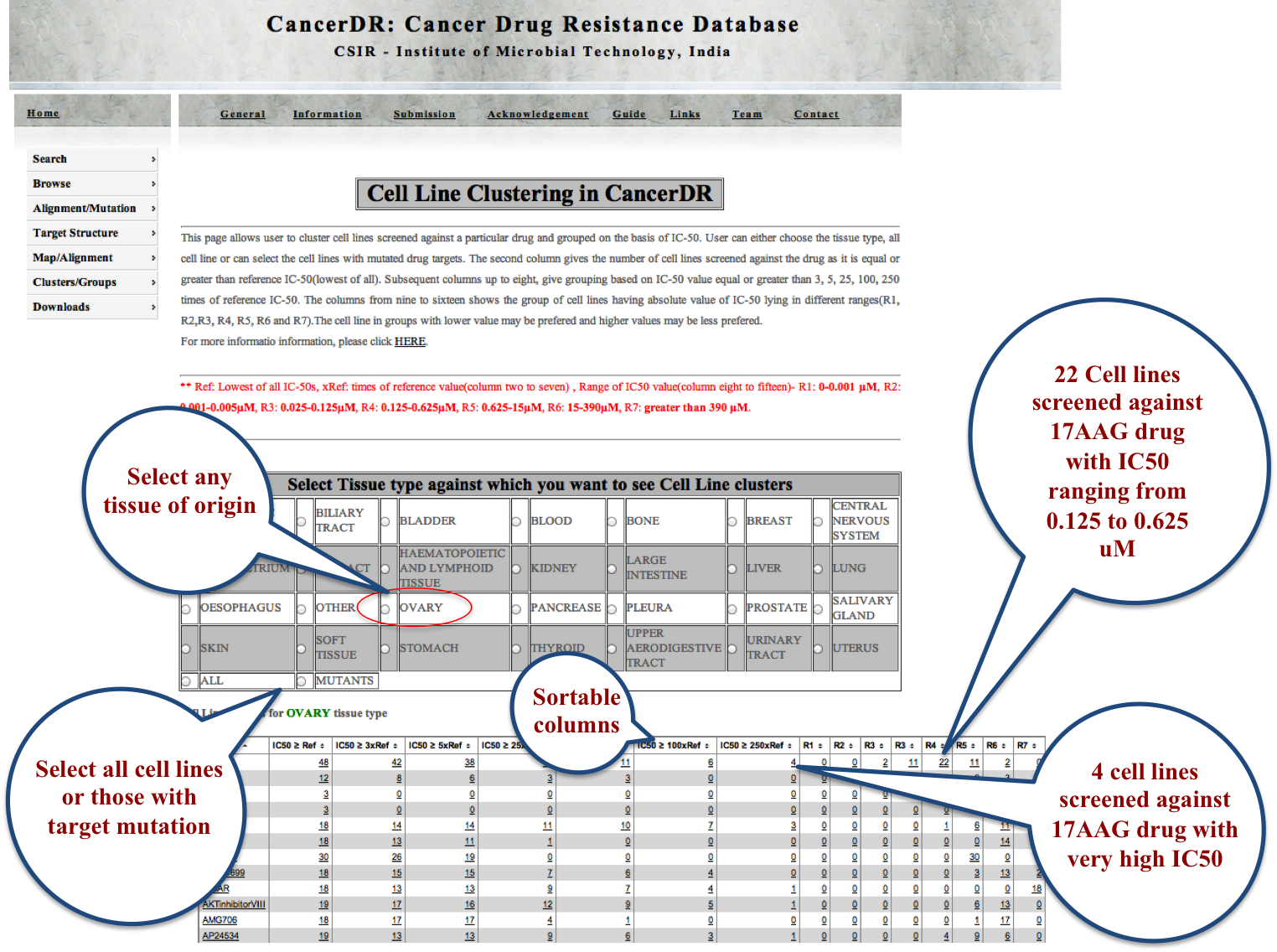 |
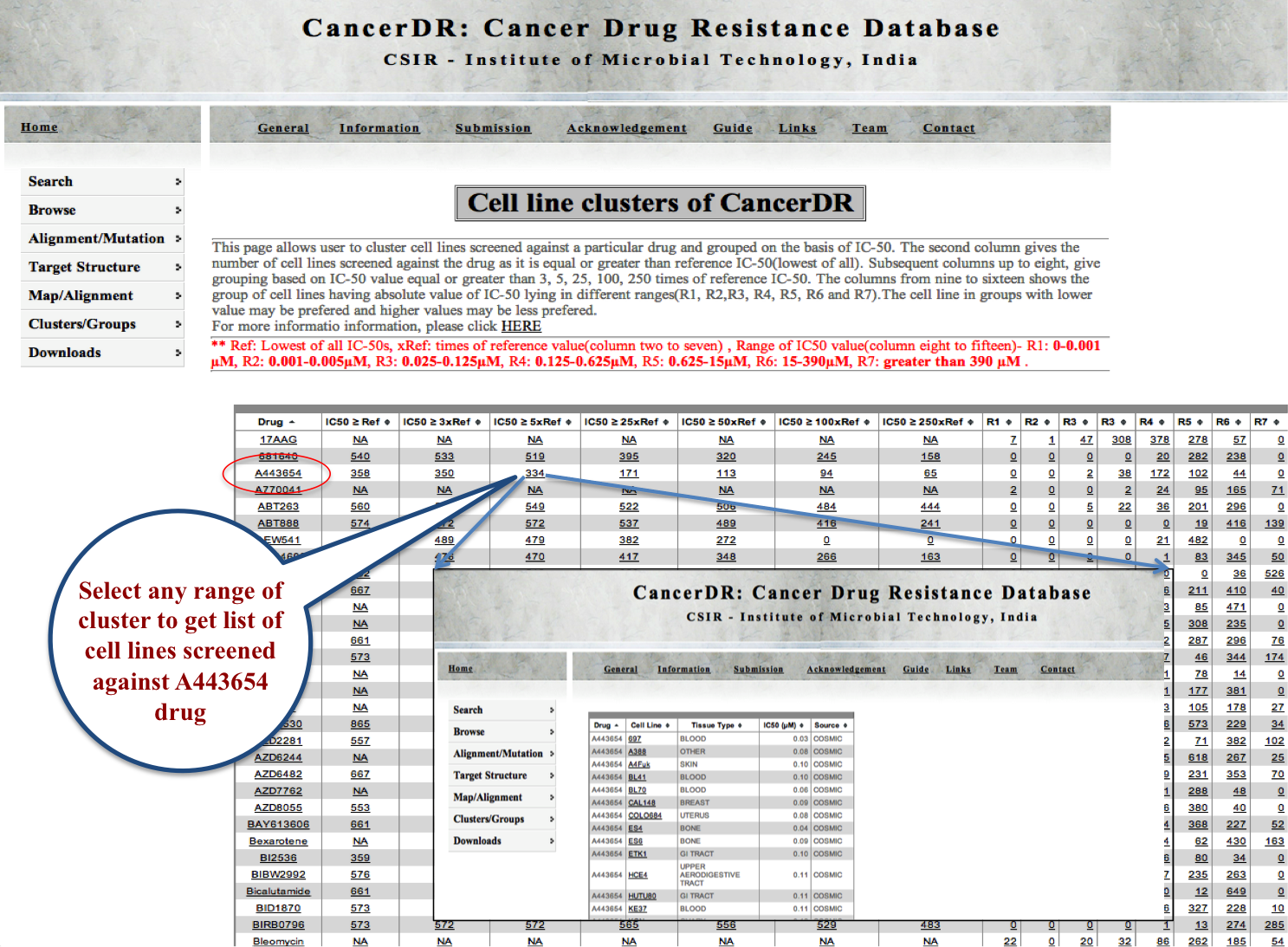 |
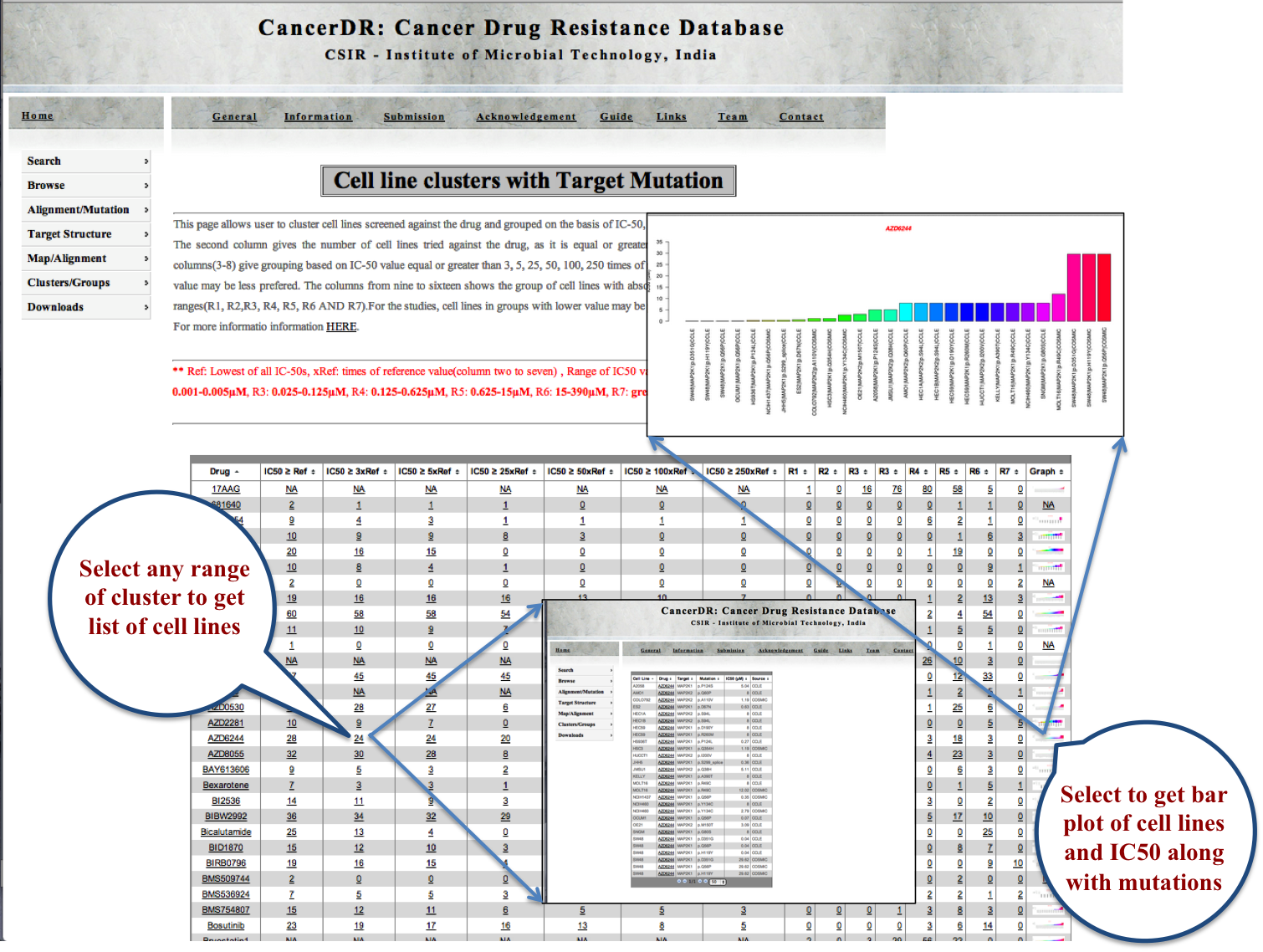 |
DRUGS This interface presents the clustering of drugs, which have been screened against any cell line. For better analysis we have given selection options for clustering of all cell lines, clustering of cell lines based on tissue of origin and clustering of cell lines for which target is mutated. The figures in columns, from second to nine, show the number of drugs, which have IC-50 in the range given in header. The ranges are given in micro molar (uM) and increasing almost logarithmically from 0.00 to 390 uM. By selecting any figures in any column say 0.001-0.005 uM in cell line row user can get group of drugs having IC-50 value lying in this range only. Lower IC-50 value groups of drugs may be preferred and drug groups having higher IC-50 value may be less preferred as they have better drug options for that particular cell line. |
DOWNLOADS |
SEQUENCES This module gives the facility to download the sequence of the drug targets along with their mutants or variant sequences in the FASTA format. User can either select a particular target and its mutants/variants or all the targets and their mutants/variants. Further, rsnyc facility is given to download all the sequence data, so that user can update data. |
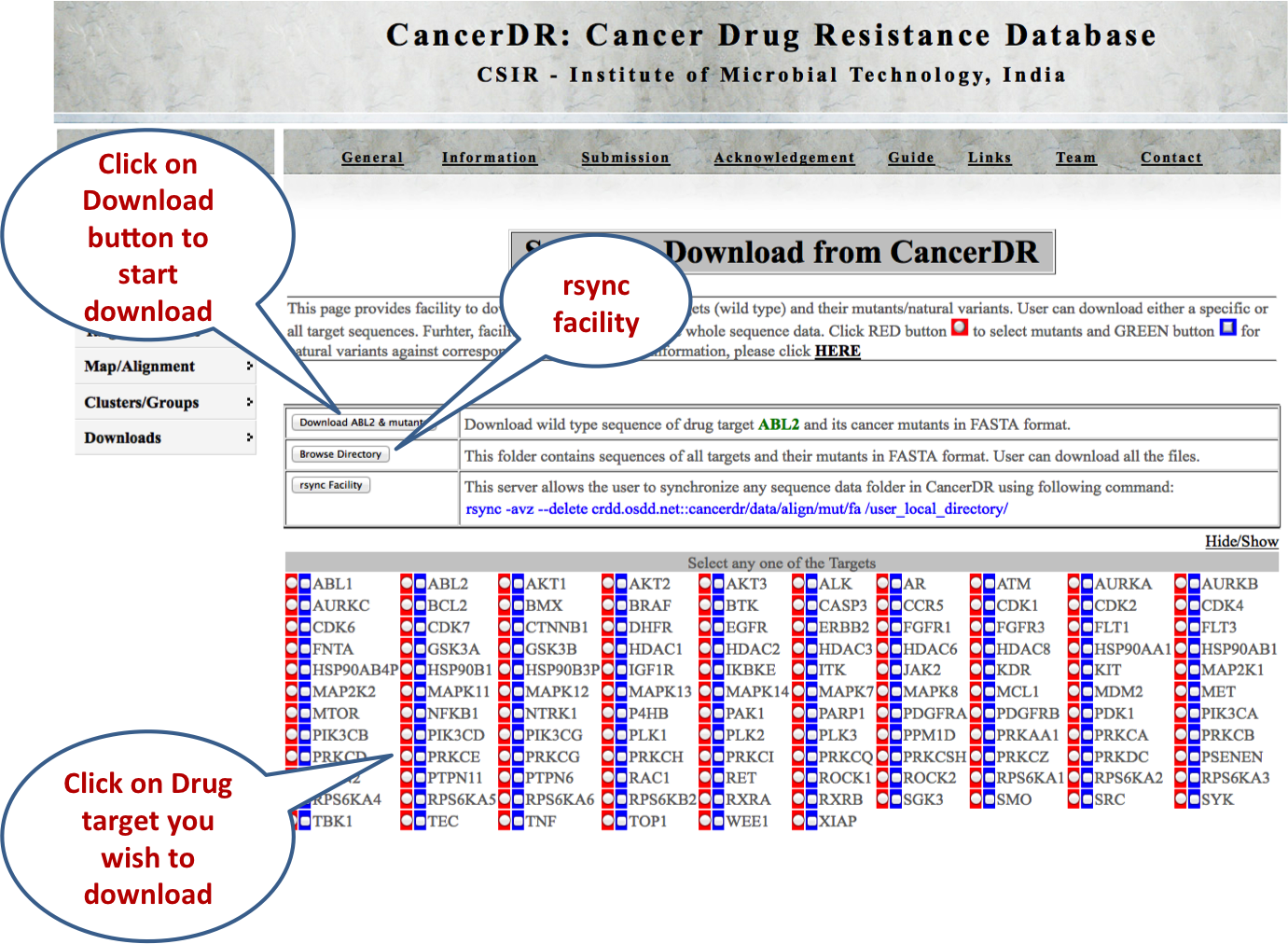 |
ALIGNMENTS This module gives the facility to download the multiple sequence alignments of the drug targets along with their mutants or variant in the CLUSTALW format. User can either select a particular target and its mutants/variants or all the targets and their mutants/variants. Further, rsnyc facility is given to download all the alignment data, so that user can have updated data. |
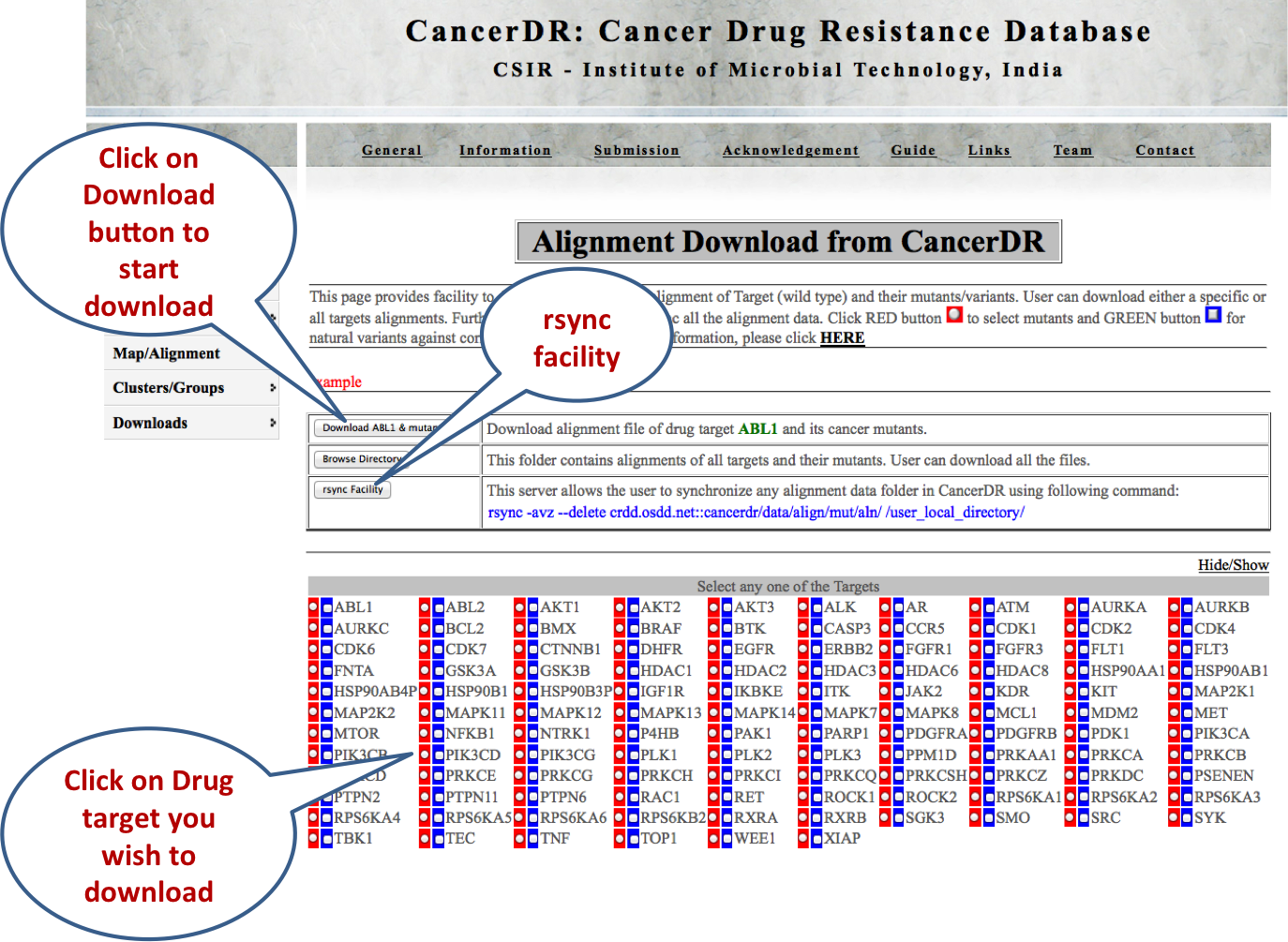 |
STRUCTURES This module gives the facility to download the predicted structures of the drug targets along with their mutants or variant sequences in the PDB format. User can either select a particular target and its mutants/variants or all the targets and their mutants/variants. Further, rsnyc facility is given to download all the structure data, so that user can have updated data. |
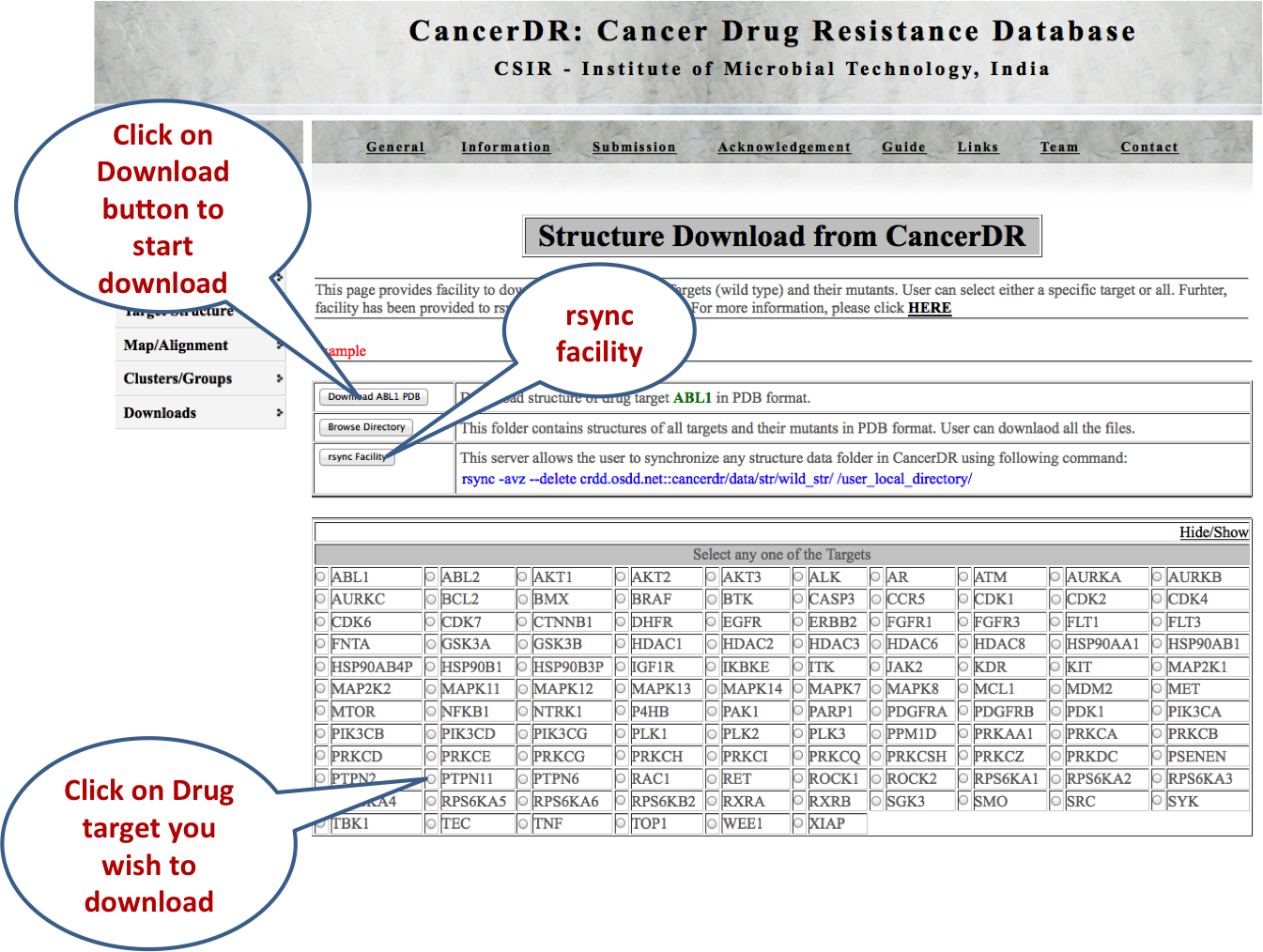 |
EXPERIMENTAL STRUCTURES This module gives the facility to download the experimental structures of the drug targets available in the PDB in the PDB format. User can either select a particular target or all the targets. Further, rsnyc facility is given to download all the structure data, so that user can have updated data. |
 |
FREQUENTLY ASKED QUESTIONS |
Q1. What is CancerDR? Ans. CancerDR is the collection and compilation of pharmacological profiling of 148 major anticancer drugs on 952 human cell lines. CancerDR data is mainly divided into two parts, first primary data which includes drug sensitivity information (i.e. IC50 value) of each anticancer drug on various cancer cell lines, information about drugs, about cell lines and the molecular targets and the mutation data of target genes. Secondary data includes prediction of tertiary structures of the targets, sequence alignments, structural alignments, clustering of cell lines and drugs etc.
Q6. Is CancerDR data downloadable? |
