General information:-
For a peptide to be a T-cell epitope, it is necessary that it must be a MHC binder. In th past much work has been done in developing methods for prediction of MHC binders and non-binders. But along with this information, one is always anxious to find out the binding affinity of these peptides for the MHC molecules. So keeping this demand of the immunologists in mind we got inspired to develop a SVM based method to predict the binding affinity (natural logarithm of IC50 values) of the peptides.
This method uses either the binary pattern and or the amino acid composition of the amino acids for making the predictions.
Stepwise help:-
Name of protein: This is an optional field.The name of the protein may have letters and numbers with the "-" or "_". All other characters are non-permissible. The field is assigned a default name "Protein".
The sequence name is just used only for your information. It may be a problem with ? , for example or an empty space within the name of the sequence, which is not allowed for reasons of security.
Protein sequence:- This server allows the submission of the sequence in any of the standard formats. The user can paste plain sequence in the provided inbox. The server also has the
facility for uploading the local sequence files. Amino acid sequences must be entered in the one-letter code. All the non standard characters will be ignored from the sequence.
Sequence format: The server can accept both the formatted or unformatted raw antigenic sequences. The server uses ReadSeq routine to parse the input. The user should choose whether
the sequence uploaded or pasted is plain or formatted before running prediction. The result of the prediction will be wrong if the format chosen is wrong.
Prediction Approach:
1.Binary pattern of amino acids:- If a user selects this prediction approach then the binary pattern of the query sequence will be provided as input to the method and a model devloped by using the binary pattern of the trainingset as input, will be used for making predictions. (For detailed information please visit the algorithm page)
2.Amino acid composition:- Selecting this prediction approach will use a model that is developed by training the method on the amino acid composition of the trainingset. Also the amino acid composition of the query sequence will be provided as the input to the prediction method.
Threshold:A threshold value is used to discriminate binders from non-binders. User can enter a IC50 value in nM as threshold. Pepties having a predicted natural log of IC50 value less than or equal to the threahold are the predicted HLA-DRB1*0401 binders (because lower the IC50 higher is the affinity) and those having values greater are the predicted non-binders.
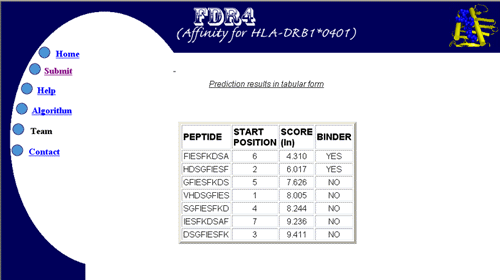
Prediction result:- After the analysis the result having nonamer subsequences of the protein, and their predicted affinity in terms of natural log of IC50 value can be illustrated both in graphical or in tabular form. A sample of these is shown below.
1.Display in graphical form: In this form of diplay all the predicted binding cores are diplayed in seperate lines.This diplay provides a clear indication of the position of predicted. An example is shown below-

1. display in tabular form The nonamer subsequences are displayed in a table in ascending order of their score (because loewr IC50 indicates higher affinity). The server also provides the facility to customize the number of best binders to be displayed in the table.e.g.