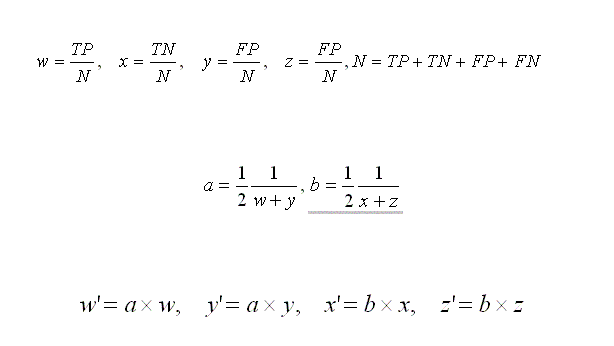|
Introduction:
The performance of different gene-finders were evaluated on the accuracy measures defined below. The various measures have been taken as defined in Burset and Guigo, 1996 and Rogic et al, 2001.
- NUCLEOTIDE LEVEL ACCURACY:
- True Positives (TP):
The number of nucleotide bases that are actually coding and are predicted as coding.
- True Negatives (TN):
The number of nucleotide bases that are actually non-coding and are predicted as non-coding.
- False Positives (FP):
The number of nucleotide bases that are actually non-coding and are predicted as coding.
- False Negatives (FN):
The number of nucleotide bases that are actually coding and are predicted as non-coding.
- Sensitivity (Sn):
 . .
- Specificity (Sp):
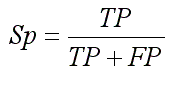 . .
- Correlation Coefficient (CC):
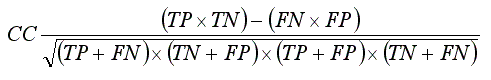 . .
- Simple Matching Coefficient (SMC):
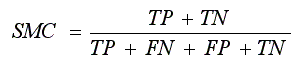 . .
- Average Conditional Probability (ACP):
 . .
- Approximate Correlation (AC):
 . .
- EXON LEVEL ACCURACY:
- Sensitivity (ESn):
 . .
- Specificity (ESp):
 . .
- Average (EAvg):
 . .
- Correct Exons (CR):
Proportion of predicted exons whose both ends are correct.
- Partially Correct Exons (PC):
Proportion of predicted exons whose either 5' or 3' alone is correct.
- Overlapping Exons (OL):
Proportion of predicted exons whose end are incorrectly predicted but overlaps an actual exon.
- Missed Exons (ME):
Proportion of actual exons which do not overlap any of the predicted exons.
- Wrong Exons (WE):
Proportion of predicted exons which do not overlap any of the actual exons.
- PROTEIN LEVEL ACCURACY:
- Sensitivity (Psen):
Ratio of number of predicted protein-coding genes whose entire protein translation product correctly matches the actual protein product of the corresponding genes against number of actual protein-coding genes.
- Specificity (Pspe):
Ratio of number of protein-coding genes correctly predicted by the programs against total number of predicted protein-coding genes.
|