Help |
|
MtbVeb has been developed to address the vaccine issue for Mycobacterium tuberculosis in every possible direction for a new vaccine. We have attempted to find a novel vaccine strain specific to virulent strain of interest, analysing proteins may be used in recombinant BCG or subunit vaccine and peptide based vaccine from predicted immune response. Beside predicted immune response analysis, we have also mapped already known sequence for a particular immunity. These known sequences were extracted and mapped from Immune Epitope Database (IEDB).
|
|
The complete architecture of Mtbveb will be descirbed under following sub-headings: |
|
|
|
|
|
1. Strain-specific Vaccine In the strain-specific page, we have provided the user to browse strain-wide experimental epitopes, genome visualization, strain-specific vaccines and tool to predict vaccine candidates for user-defined strains  |
|
1.A Browse strain This page will help user to browse experimental epitopes mapped from IEDB to each sequence present in that particular bacteria. Here, the java-applet based graphical represnetation will visualize the Bcell , T cell and MHC binding assay comparatively in each strain or total assay will be shown on default mode. Besides the number of assays, the percentage value for each assay category is given in graphs-legends.  |
If the user choose to browse particular category of assay for the strain of interest. The outcome will be presented in tabular format, where each epitope is shown with host and source information.
 |
|
1.B Compare antigens We have compared 4003 proteins of Mtb H37Rv with the proteome of each strain and selected top hit based on global alignement generated using EMBOSS package needleall program. The top hit from each strain is extracted and aligned with corresponding Mtb H37Rv protein using clustalw. The alignment file can be visulaized usign java applet. 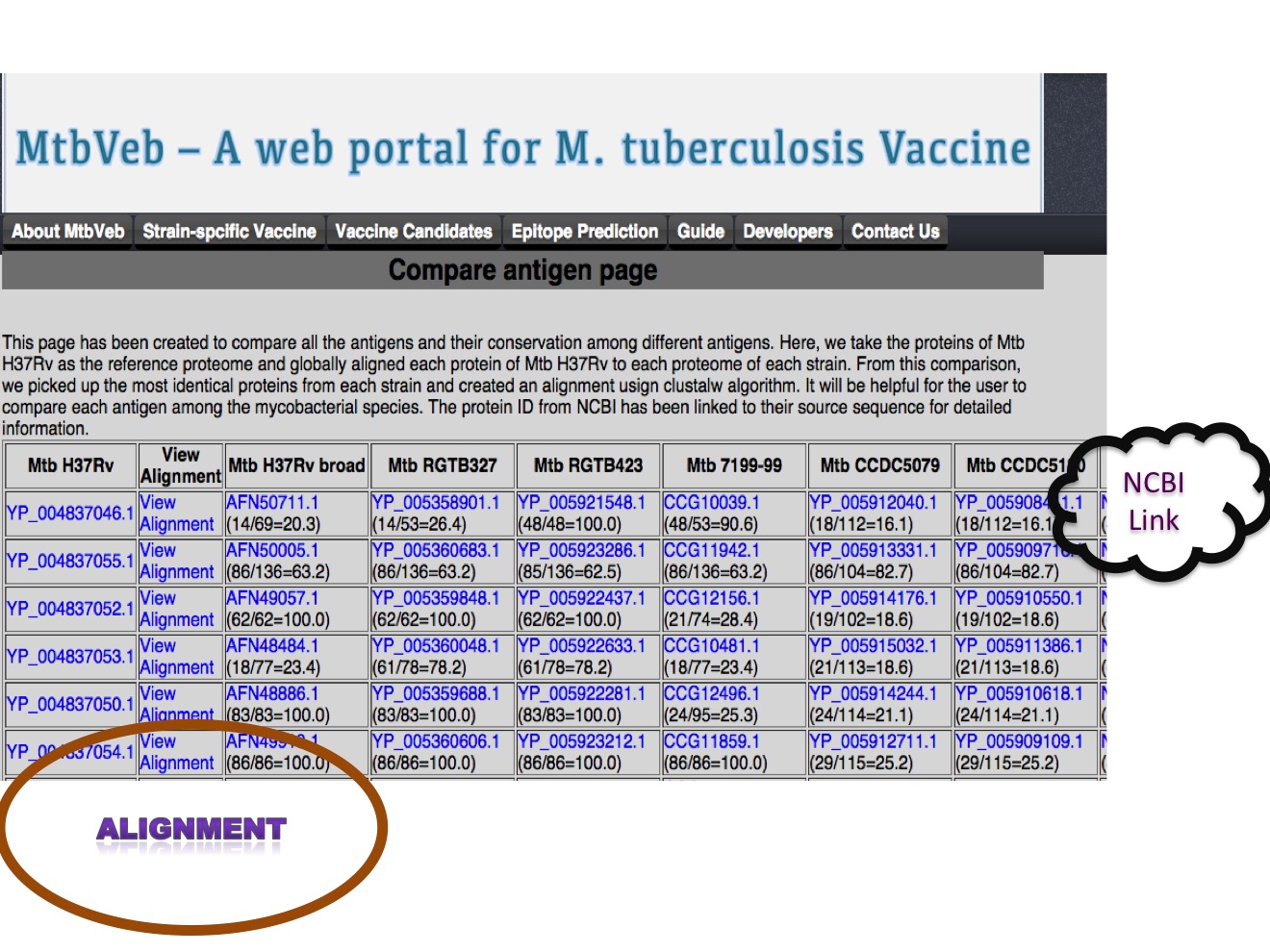 |
The alignment file has been generated for each proteins using clustalw. The alignment file can be visualized using Jalview web applet.
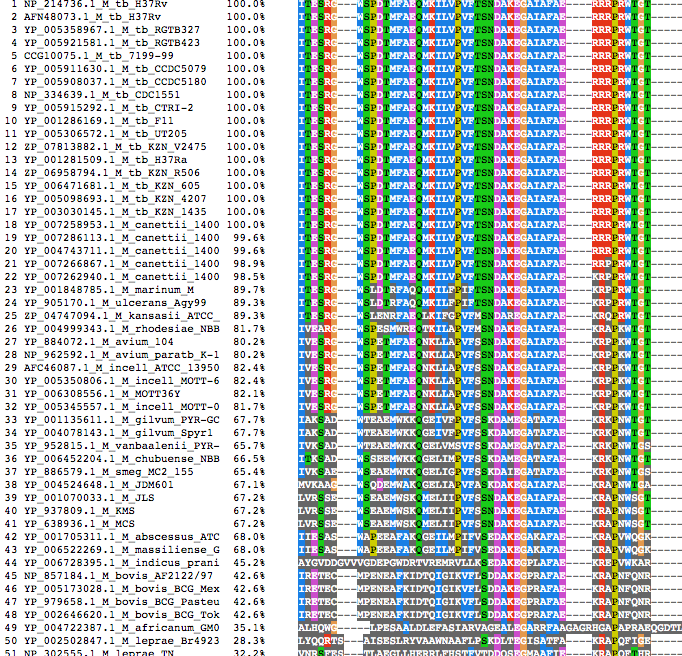 |
|
1.C Vaccine strain In this module different antigens (178 in our study) were presented, in a way where user can have idea how many number of proteins were found in strain in reference to Mtb H37Rv. There is three table, one table for each category of proteins from virulent, secretion and region of genetic variance. 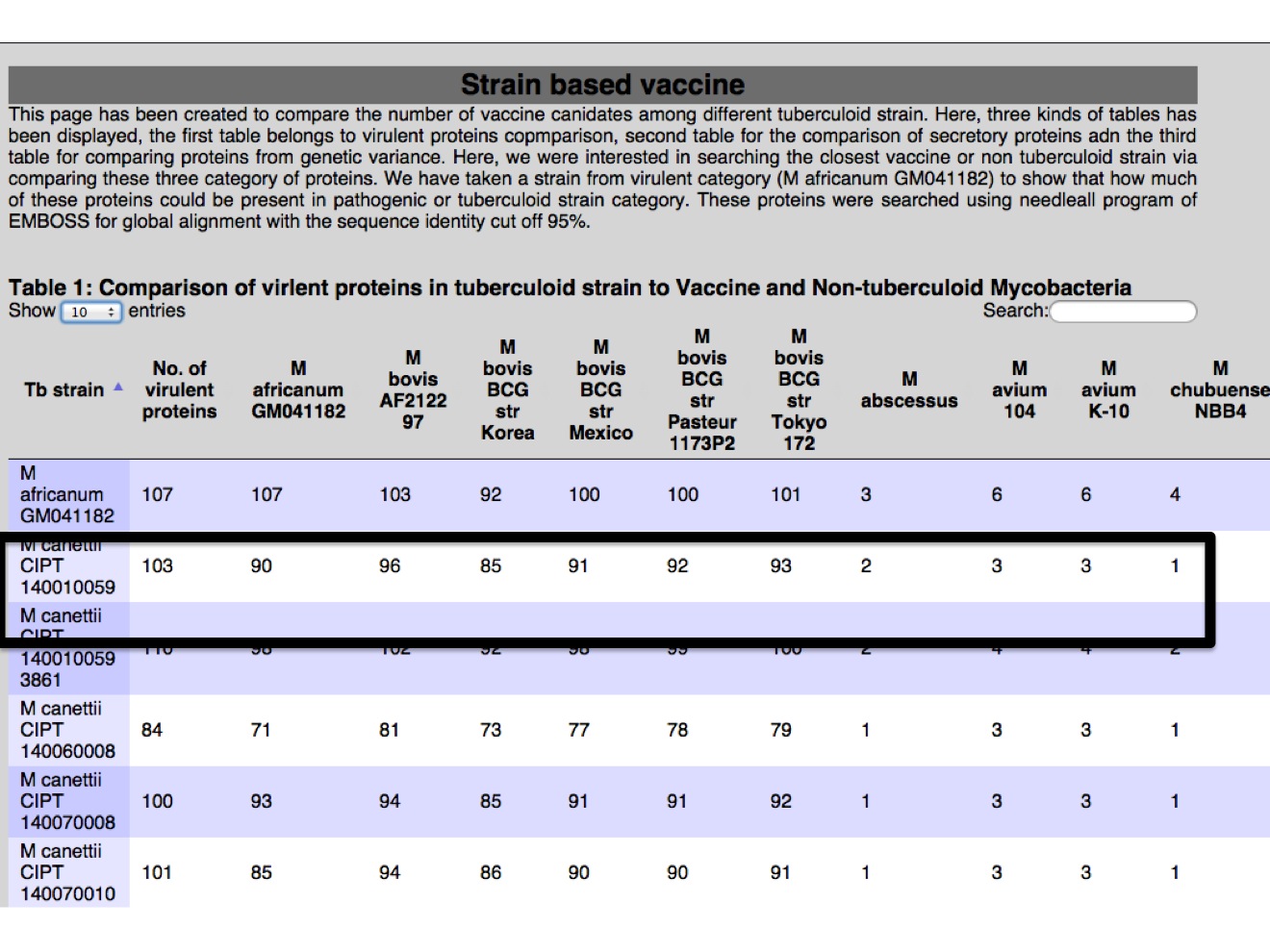 |
1.D New strain
We have allowed user to carry out the analysis for the strain other than the strains present in our study. User can carry forward this analysis from each kind of files available with him/her.
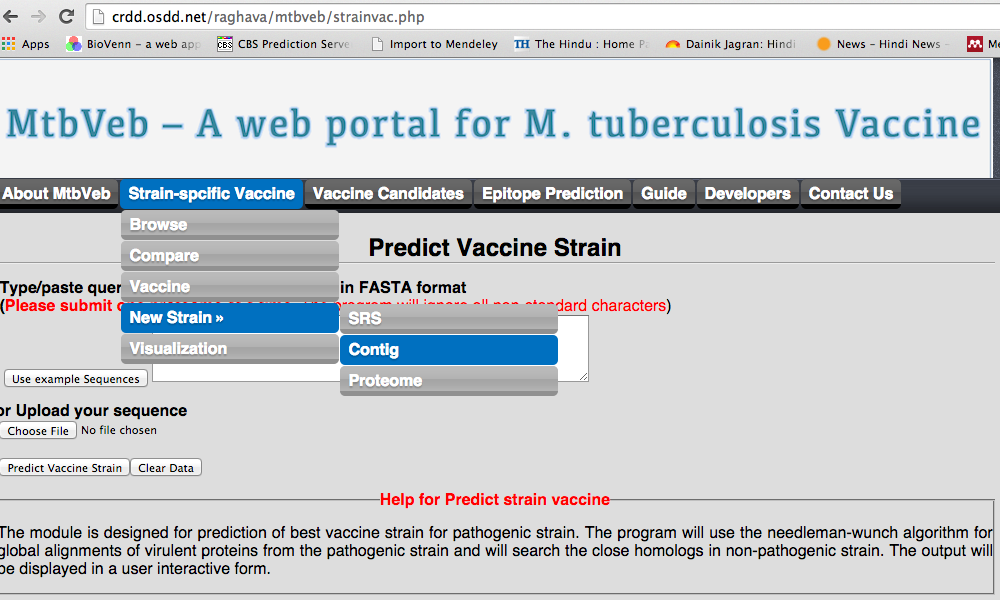 |
|
1.E Genome visualization We have implemented three kind of genome visualization tool for deep insight and beautifully presentation of the genome of each strain. Argo applet based genome visualization 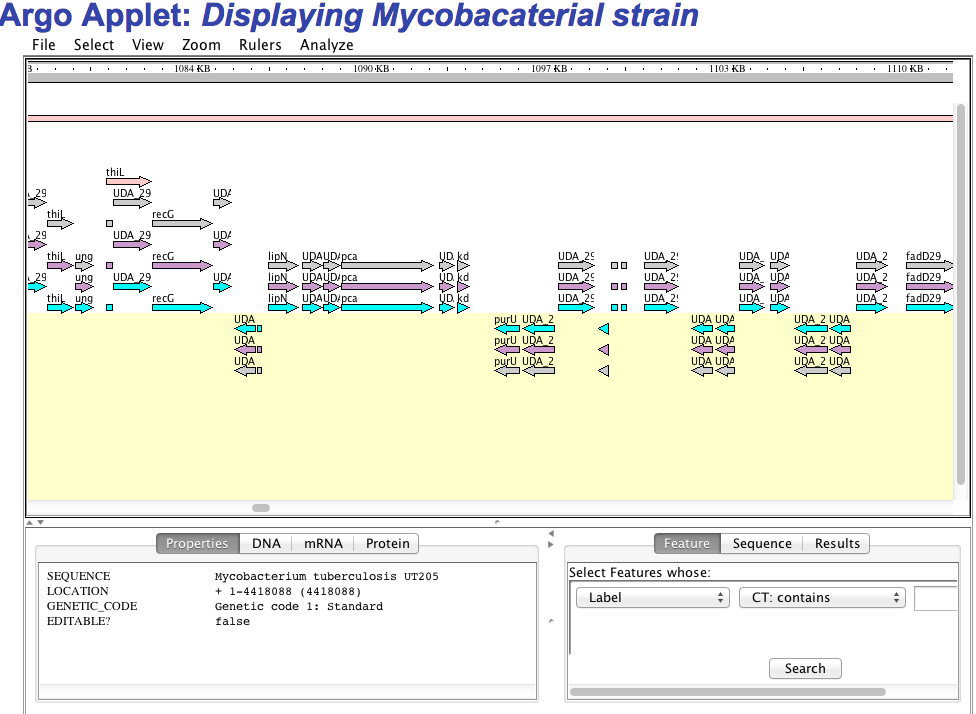 CGview applet based circular genome visualization 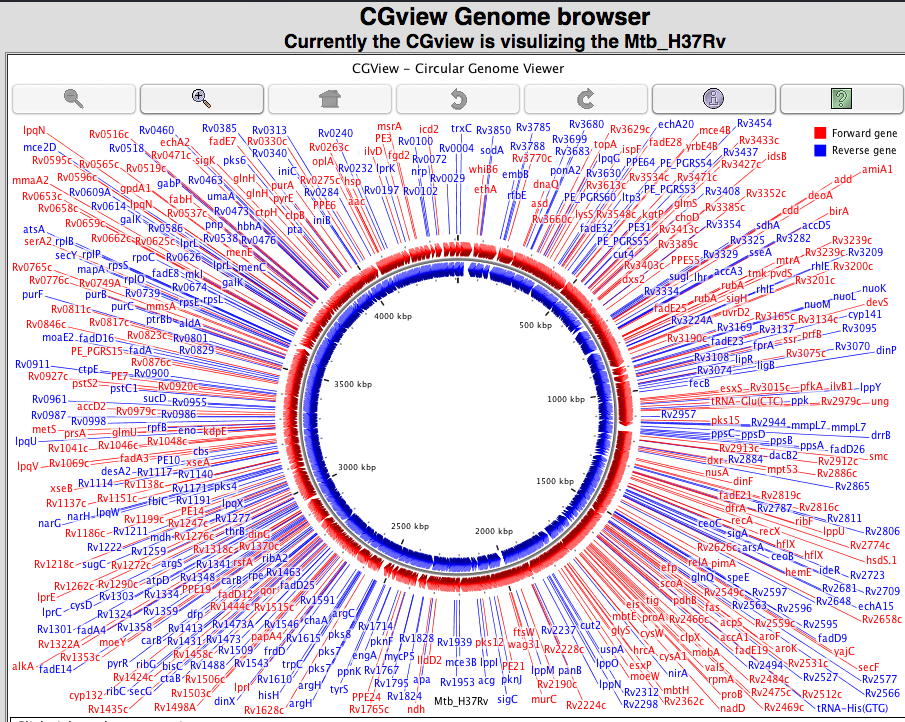 Jbrowse based genome visualization  |
|
2. Canidate vaccine The candidate vaccine were decided from three category of proteins, virulent, secretory and region of genetic varaince  |
|
2A. Targets We are providing a table for each category and each table have displayed for the presence or abscence of particular antigen in all strains from our study.  |
|
2B. Antigen compare This tool has been provided to compare the protein of user-interest with all the mycobacterial strain and human proteome. 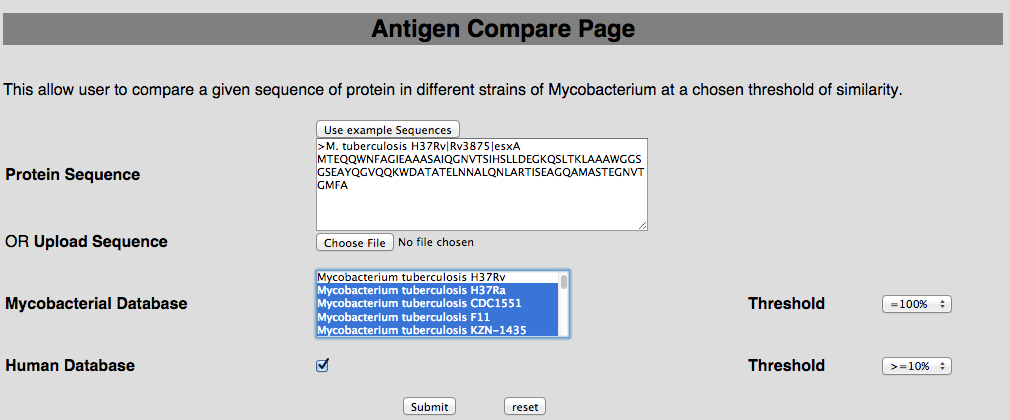 |
|
2C. Potential vaccine candidates The potential vaccien canidates were selected based on the comparison of protein in different category of strain and thier known immune response. 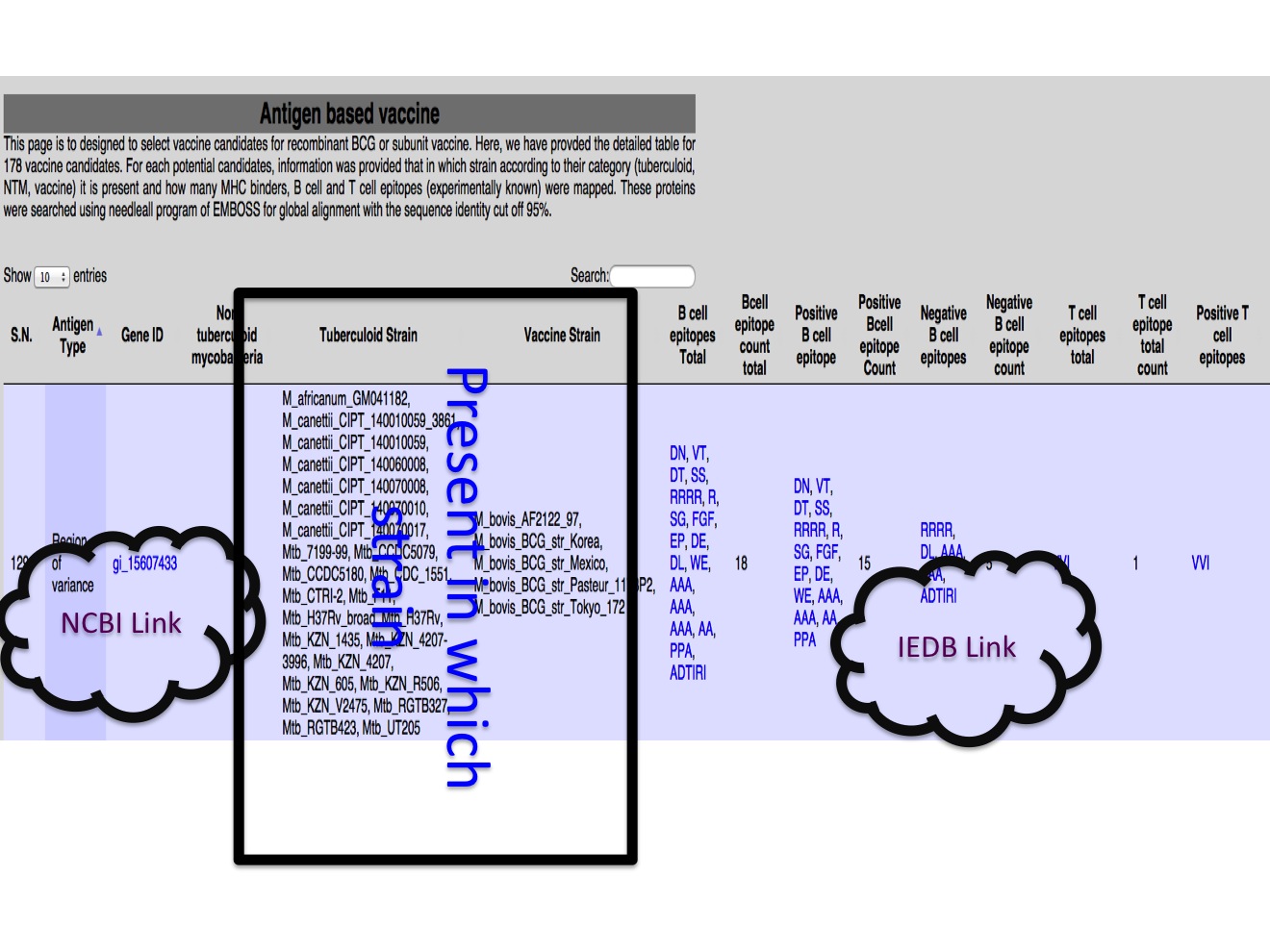 |
|
2D. Alignment of candidates The alignment file has been generated for each proteins using clustalw. The alignment file can be visualized using Jalview web applet.  |
|
2E. Map epitope on antigen This tool has been provided to map the epitope in the protein of user-interest. In mapping we match epitope of IEDB with peptides generated with user-selected window length. The exactly matched epitopes were displayed in tabular format. 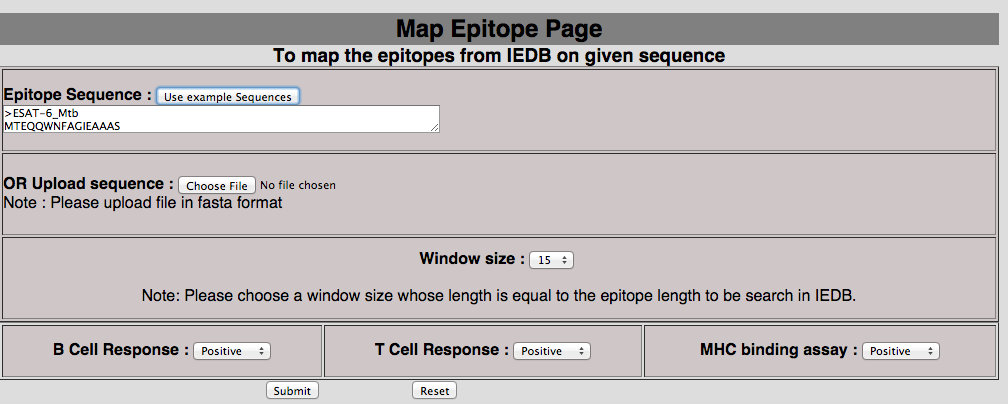 |
|
3. Epitope prediction baesd vaccine Here, we have analyzed all the peptides created using fixed window length of 9 size. Here user can browse, search and predict the peptides for different type of immune response.  |
|
3A. Browse epitope In this browse option, user can extract the peptide of desired immune response. In the example, we have browsed the peptide that were predicted to be showing Th1 kind of immune response. Th1 has been defined for binding to MHC class II (predicted using propred) and inducing interferon-gamma response (predicted using IFNepitope server).  |
|
3B. Advance Search This module has been developed to filter the peptide with desired logical filters. This will help the user in finding the peptide with his/her interest from their own defined filters. 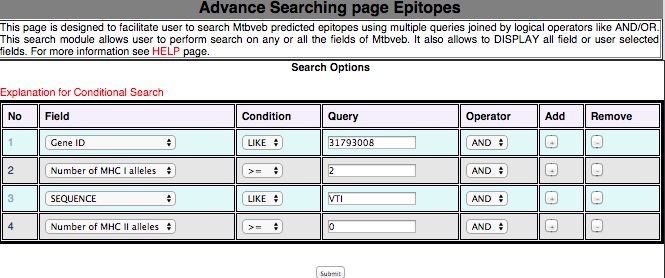 |
|
3C. Peptide based vaccine This page displays the peptide that were predicted to be MHC class-I binder (predictes using Propred1), MHC class-II binders (predicted using Propred), CTL epitopes (predicted using CTLpred and nhlapred) and Th1 epitopes (predicted using IFNepitope server). We hope this peptide will be potential vaccine candidates. 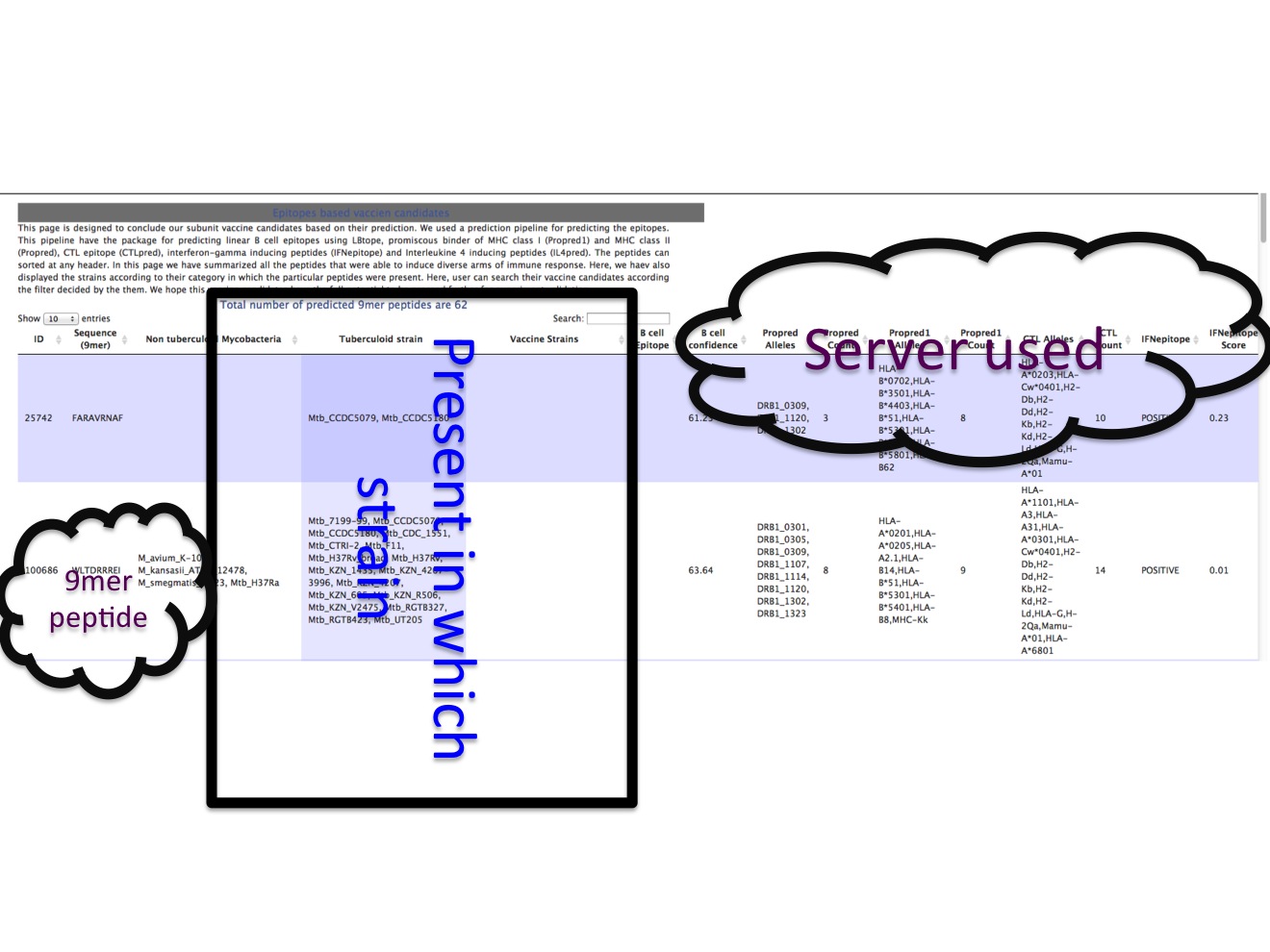 |
|
3D. Prediction of user-peptides This module will be very helpful for user to predict the immune response from our pipeline. Here, we have implemented it for user to generate all possible 9mers and predict immune response with Propred1, Prored, CTLpred, nHLApred, IFNepitope and IL4pred. 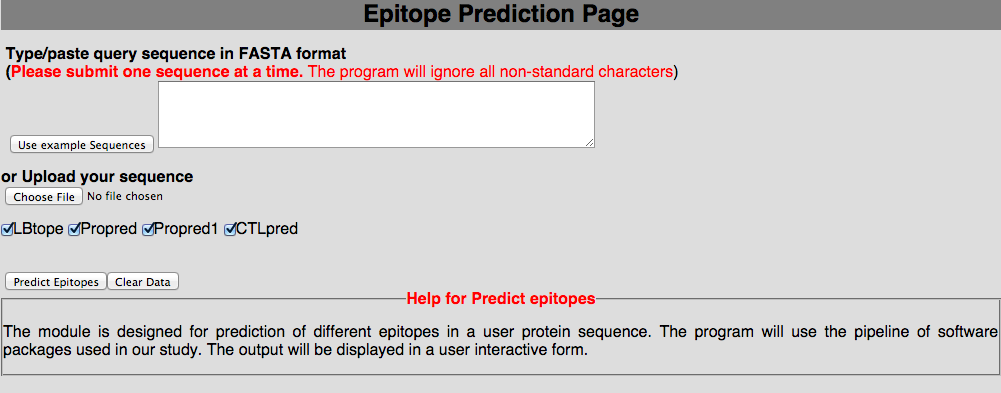 |