ToxinPred is an in silico method, which is developed to predict and design toxic/non-toxic peptides. The main dataset used in this method consists of 1805 toxic peptides (<=35 residues).
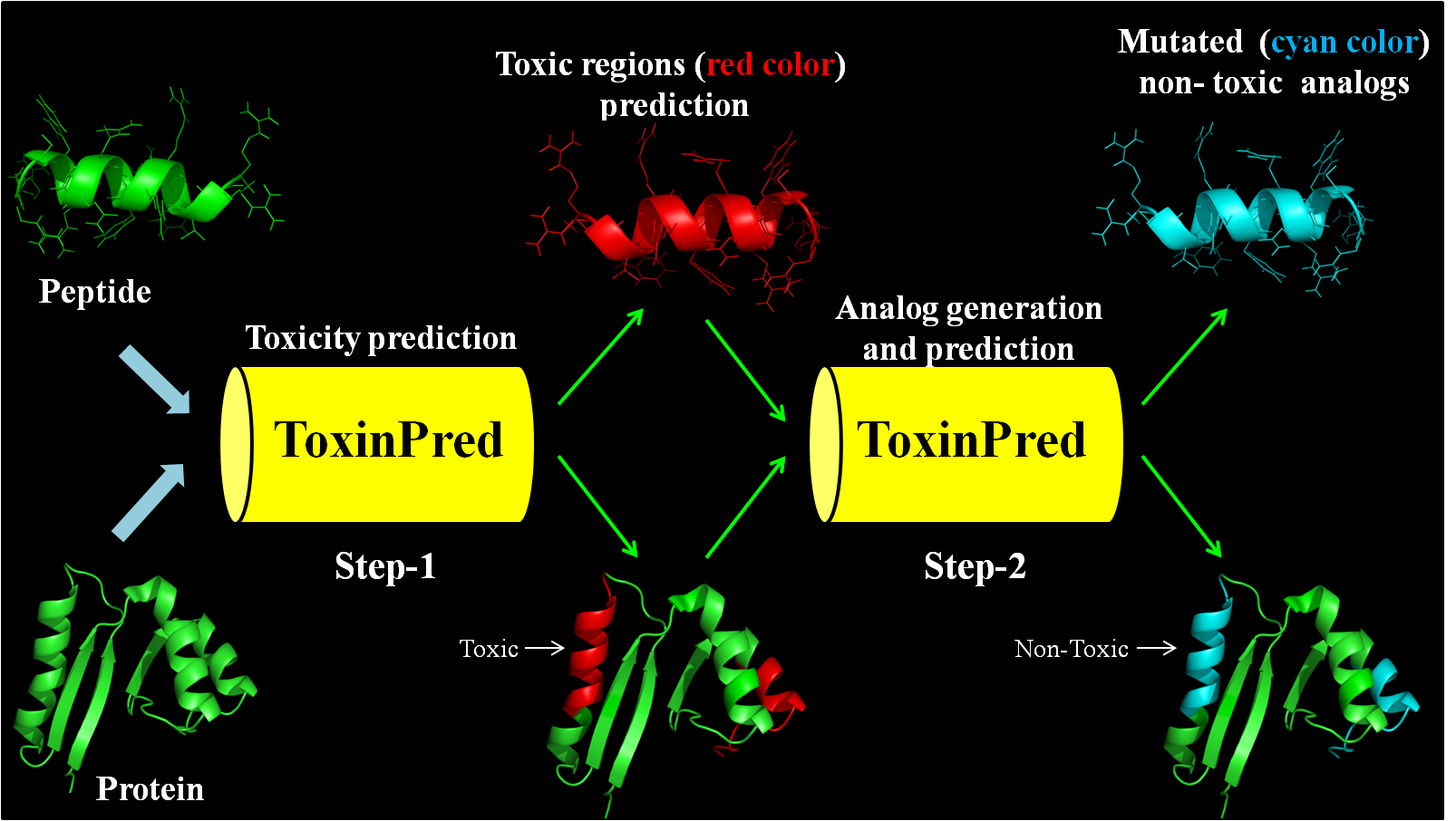
(1) Designing Peptide: This module allows user to generate all possible single mutant analogs of their peptides and predict whether the analog is toxic or not.
(2) Batch Submission: This module of ToxinPred allows user to predict number of toxic peptides submitted by the user.
(3) Protein Scanning: This module generates all possible overlapping peptides and their single mutant analogs of protein submitted by the user. It also predicts whether overlapping peptide/analog is toxic or not.
(4) QMS Calculator: This tool allows the users to submit query peptide in FASTA format and to optimize the peptide sequence to get maximum/minimum/desired toxicity based upon the Quantitative Matrix based position specific scores. It will help the user to tweak any residue from the predecessor peptide to attain the analog with desired property (highest/lowest toxicity).
One of the major features of server is that it also calculates various physicochemical properties. Peptide analogs can be displayed in sorting order based upon desired properties.
| Reference: Gupta et. al.: In silico Approach for Predicting Toxicity of Peptides and Proteins. PLoS ONE 8(9):e73957. doi:10.1371/journal.pone.0073957 Link |