Algorithm
Antiviral compounds
Antiviral compounds are a category of antimicrobial drugs used specially for treating viral infections by inhibiting the development of the viral pathogen inside the host cell. A growing number of antiviral drugs are designed to help deal with HIV, HCV, HHV, HBV and other viruses etc. Researchers are working to discover new antivirals (natural/synthetic) to extend the range of these compounds as well as to address virus resistance.
Datasets
We used the data from the ChEMBL (https://www.ebi.ac.uk/chembl/target/browser/). In this project,we selected HIV, HCV, HHV, HBV (Virus specific datasets) and 26 other viruses (General dataset) targeted by 389, 467, 112, 124 and 1391 compounds respectively.
Chemical Features
To develop QSAR models, we have calculated the different chemical descriptors (1D, 2D and 3D) and fingerprints by using PaDEL software (http://padel.nus.edu.sg/software/padeldescriptor/). These relevant chemical descriptors were extracted using attribute selection algorithms viz. 'RemoveUseless' filter followed by ClassifierSubsetEval (attribute evaluator) with BestFirst (search method) module available in Weka package.
QSAR Models
We have developed separate QSAR models for each of the 4 viruses (HIV, HCV, HHV, HBV) by using SMOreg algorithm available in Weka machine learning package. SMOreg implements the support vector machine in regression mode. Selected molecular descriptors calculated by the PaDEL were used as input.
Cross Validation
All models were evaluated using ten-fold cross validation technique as well as on independent test datasets. In order to evaluate performance of our models, we used Pearson’s correlation coefficient (R).
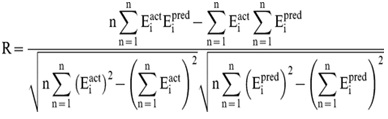
Where n is the size of test set, Eipred and Eiact is the predicted and actual efficacy respectively.

 Acknowledgements:
Acknowledgements: