How to use
The QSAR models have been integrated into a freely available and easy to use web server, 'AVCpred', where users can predict the antiviral potential of their query molecules against the different viruses in terms of percent inhibition value. AVCpred web server includes the following modules:
Submission
This allows users to submit one or more molecules at a time. Users have to choose the viruses on which they want to test their query chemical compounds. On submission, it returns with percent inhibition values against the selected viruses. Also users can view the different properties of the query molecule.
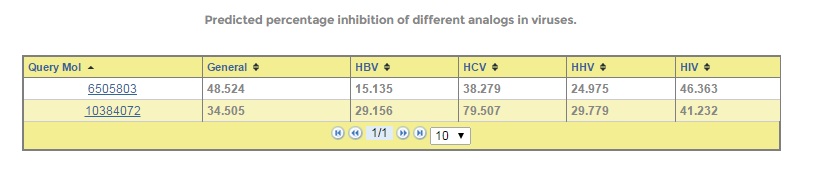
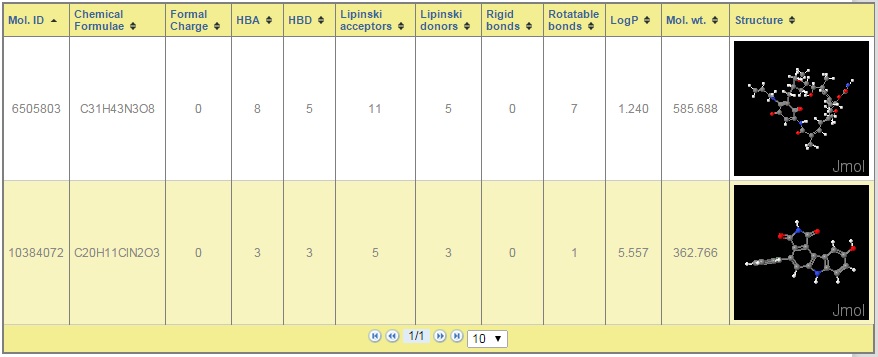
Figure: AVCpred submission form with output.
Design Analogs
It has been found that analogs of known chemical compounds are sometimes more effective than the parent molecule. In order to identify potent analogs of an existing antiviral, structural, we have included the 'Design analogs' tool, where user can design analogs based on given building blocks and predict their inhibition on the viruses.

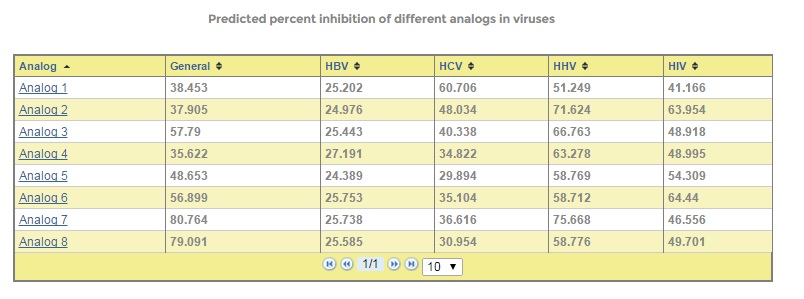
Figure: AVCpred design analogs form and output.
Draw Structure
Using the 'Draw tool' one can sketch the structure of a query molecule using JME editor. This tool also gives the predicted percent inhibition values against the different viruses In addition, one can view the various properties of the query structure.
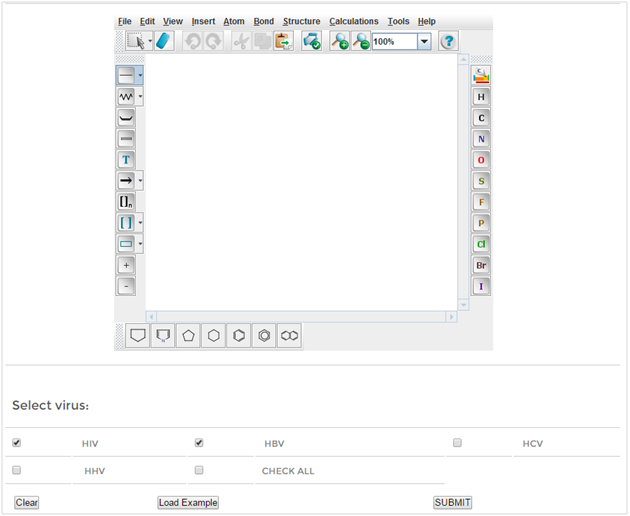

Figure: Web interface of 'AVCpred Draw' tool and result.
You can also paste the SMILES directly as shown below:


 Acknowledgements:
Acknowledgements: