Database of HIV inhibiting peptides
CONTENTS
Database | Browse | SearchAdvanced Search | Data organization | Tools | Update
HIPdb is a manually curated database of experimentally verified HIV inhibiting peptides targeting various steps or proteins involved in the life cycle of HIV like fusion, integration, reverse transcription etc. Currently the database provides experimental information of 981 natural and 87 modified peptides from varying sources tested on 35 different cell lines. Also provided are 179 reported peptides with very low/nil activity. The database provides user friendly browse, search, advanced search options and also useful services like BLAST and ‘Map’ for alignment with user provided sequences. In addition, structure and physico-chemical properties of the peptides have also been included .The database is freely available at http://crdd.osdd.net/servers/hipdb.
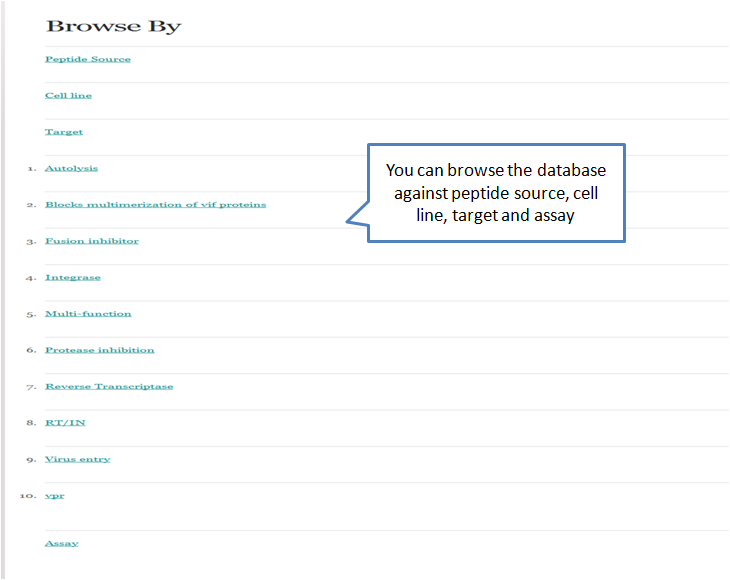
This helps the user to retrieve peptide information from database by browsing four important fields viz. peptide source, cell line, target and assay as shown in the figure.
The user can enter the desired keyword and field then select necessary fields to search the database as shown below. Here you can also download your search results as a csv file.
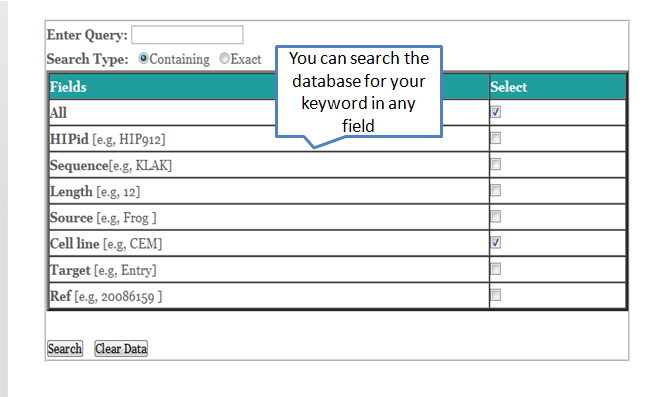
You can also make more flexible queries using 'AND/OR' etc to search the desired fields as shown below.

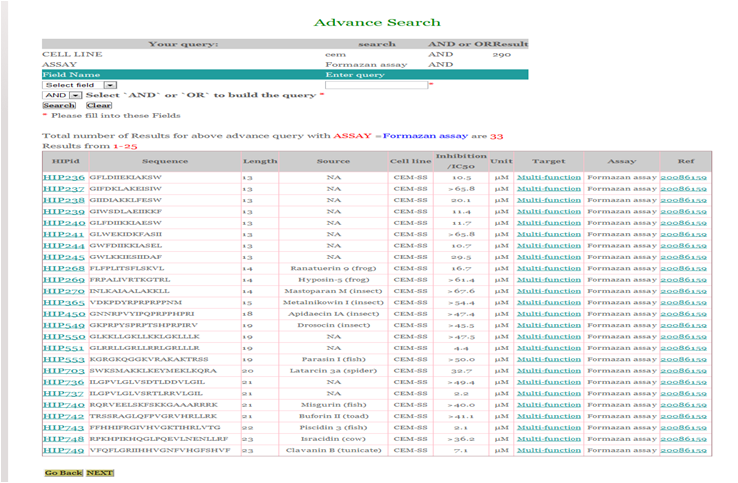
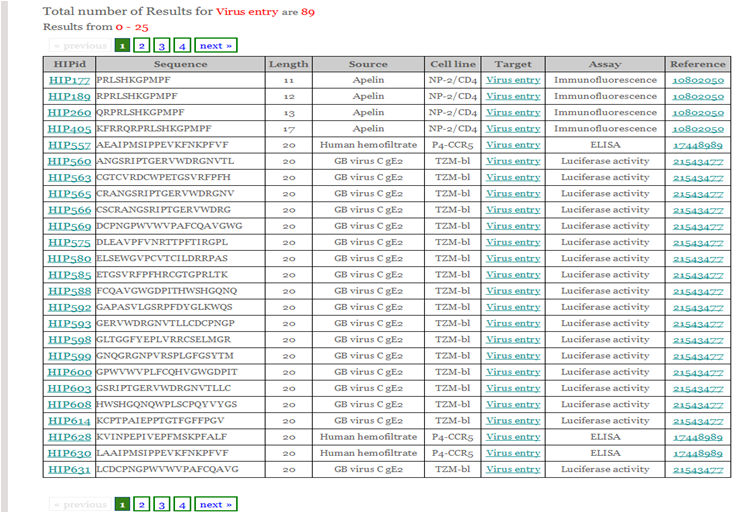
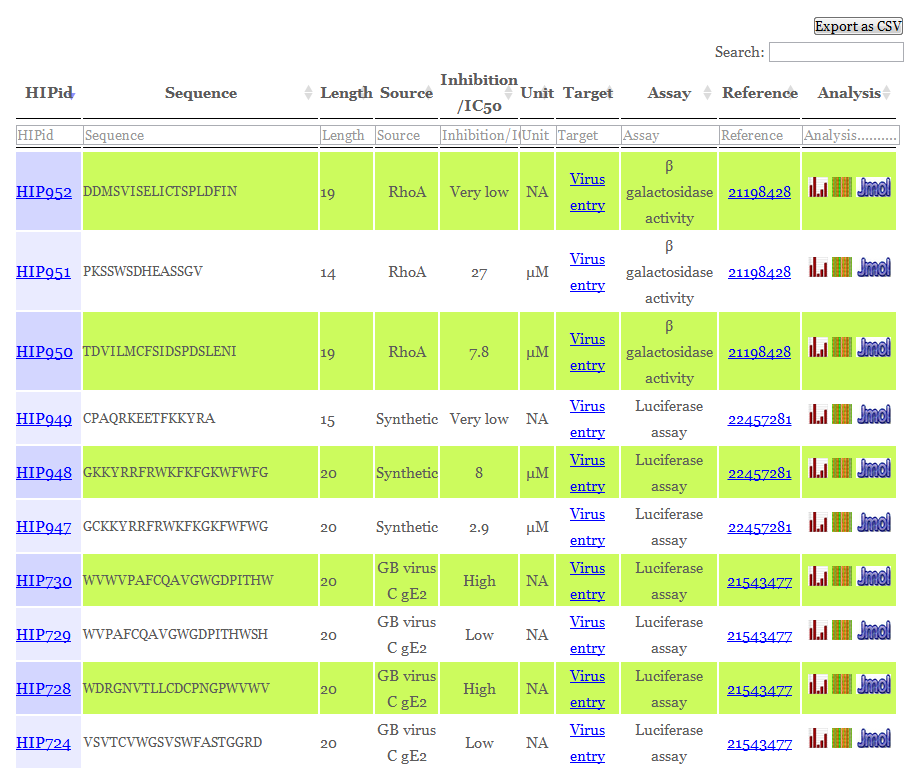
The following fields will be displayed corresponding to each record in the database

In addition to the above information two more fields including physico-chemical properties and structure of the peptide are also included in the respective record of each entry.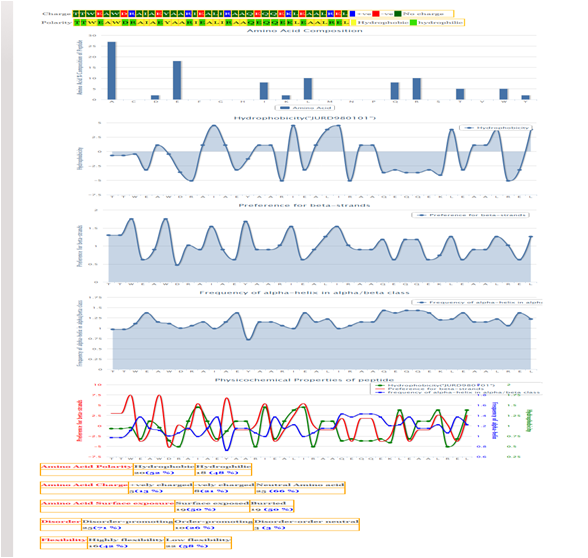
Physico-chemical properties shown are charge, polarity, composition, hydrophobicity and secondary structure preference.
Structures of the peptides were calculated using PepStr. Structures are displayed in Jmol applet.
HIPdb Map
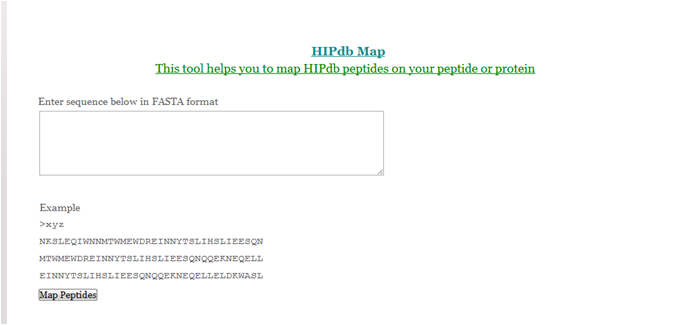

The ‘HIPdb Map’ is a simple tool to display the perfectly matching peptide available in our database to the user provided viral sequence. So, it helps the user to know that against the user provided viral sequence, how many peptides are available in our database.
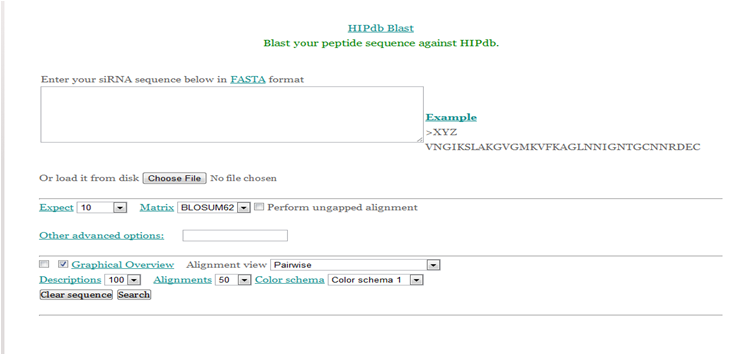
HIPdb Blast
Additionally, the Blast allows alignment of a user provided peptide sequence against all the peptide sequences available in our database. This helps the user to confirm whether a given peptide sequence or similar one has already been reported or not.

Authors generating experimental data on anti-HIV peptides are encouraged to submit the data directly into HIPdb. For this purpose, a web form for data submission is provided. Submitted information will be included in the database update after ascertaining its authenticity.
 CSIR-Institute of Microbial Technology, Chandigarh 160036, India
CSIR-Institute of Microbial Technology, Chandigarh 160036, India