Prediction of peptide antiviral activity in terms of IC50

How to use
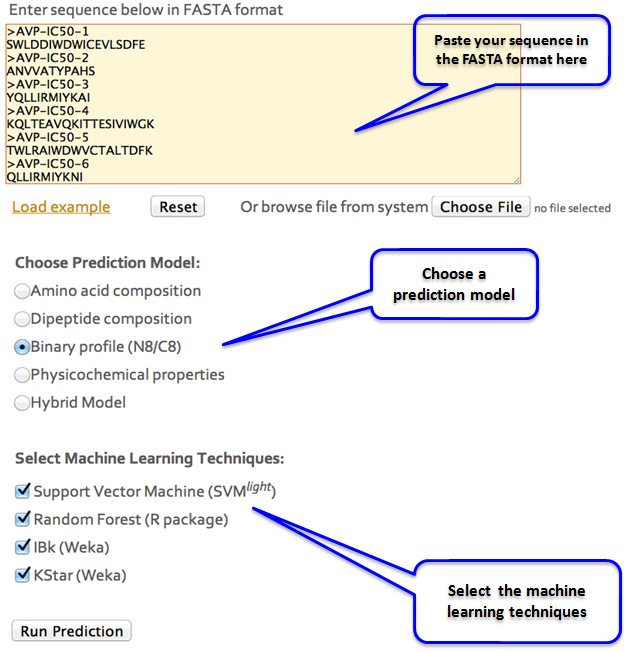
AVP-IC50Pred Input
AVP-IC50Pred submit: AVP-IC50Pred is an online, open source server to predict the IC50 values of the peptide sequences, which can be used to target human viruses. To use this server, user can simply paste the sequences in FASTA format in the text area on "Predict" button. Few examples have been provided which can be loaded by clicking on the Load example link. User can then choose the model (e.g. Amino acid composition, binary properties, physicochemical properties, Hybrid etc.) based on which the predictions will be made. Four different machine learning techniques viz. SVM (Support vector machine), RF (Random forest), IBk (Instance-based classifier.) and K* (KStar) can be selected for making predictions. User can either select any of them or keep the default settings. Clicking on Run Prediction will predict for the IC50 values of the user defined peptides.
AVP-IC50Pred Output
The prediction output is shown in tabular form with 10 columns. The first, second and third columns consist of the sequence identifier for the input FASTA sequence, the sequence itself and the sequence length respectively. The fourth column gives the action buttons for database scanning in order to check the presence of similar sequences in the existing databases (antiviral: HIPdb, AVPdb and general: Uniprot) using BLAST. Rest of the columns (5-8) is the output of different machine learning techniques used in this study (SVM, RF, IBk, K*). Each of these columns displays the predicted IC50 value for the peptide sequence in μM units. The analysis column displays the calculated physicochemical properties for the peptides in question. All the columns of the table have been provided with a sorting functionality. Search functionality is also provided to filter the results.

Result interpretation
The half maximal inhibitory concentration (IC50) is a quantitative measure which signifies the concentration of a molecule or drug required to block a given biological process by half (50%). The results in the above output may be divided into four groups, for reasons of convenience, as per the predicted IC50 values as:
Analysis Tools
Additional tools like BLAST, Map, Peptide generator and Peptide physicochemical properties calculator have been incorporated in this web server. These are briefly described as:
a) AVP-IC50Pred BLAST
AVP-IC50pred provides the user with the facility of scanning the existing databases: AVPdb, HIPdb and Uniprot to check if sequences similar to the the user provided peptides have been reported or not. The BLAST output against AVPdb is displayed as shown below. The output is similar for HIPdb and Uniprot.


b) AVP-IC50Pred Map
The MAP tool is used to fetch the perfectly matching peptides available in the existing databases which map against a user provided protein sequence. It tells as to how many peptides against the user provided protein sequence have been reported earlier.
c) Peptide Physicochemical properties
The properties tool allows the user to visually examine some important peptide features like amino acid composition, hydrophobicity, preference for β-strands and frequency of α-helix in a given peptide sequence.

d) Protein fragmentor
This tool may be used for making peptide fragments of desired length and overlapping residues from a protein sequence.

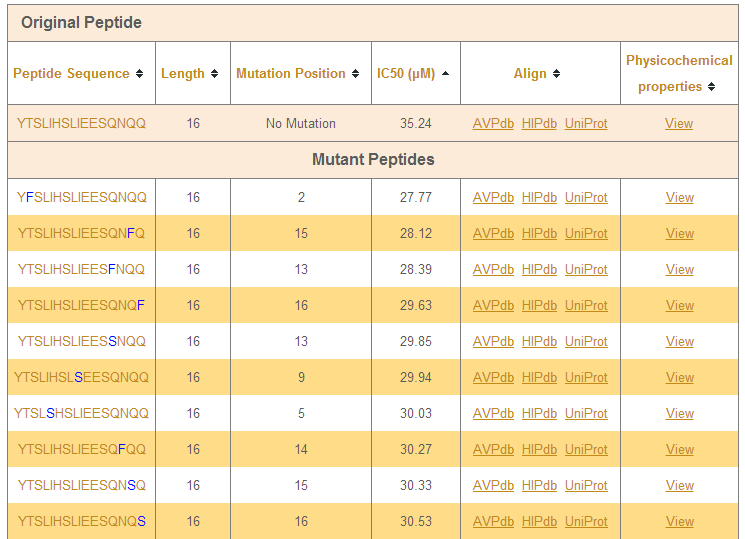
e) Mutation analyzer
This tool allows the user to generate all possible combinations of amino acid mutations in a given peptide sequence and predict the IC50 of the mutant peptides using the best performing model.
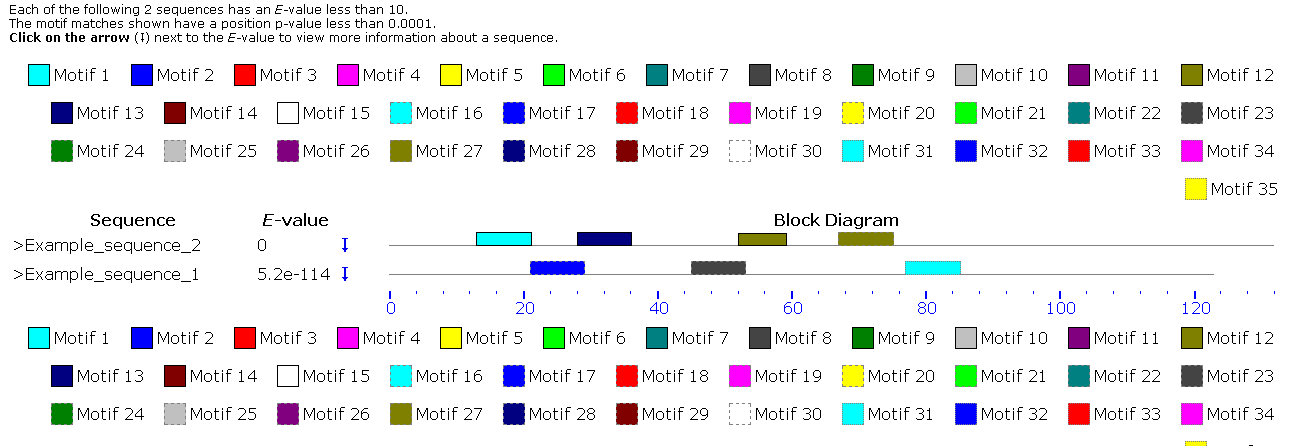
e) Motif search
AVP-IC50Pred Motif Scan allows to search possible AVP motifs in user provided protein/peptide sequences. This tool is based on MEME/MAST software
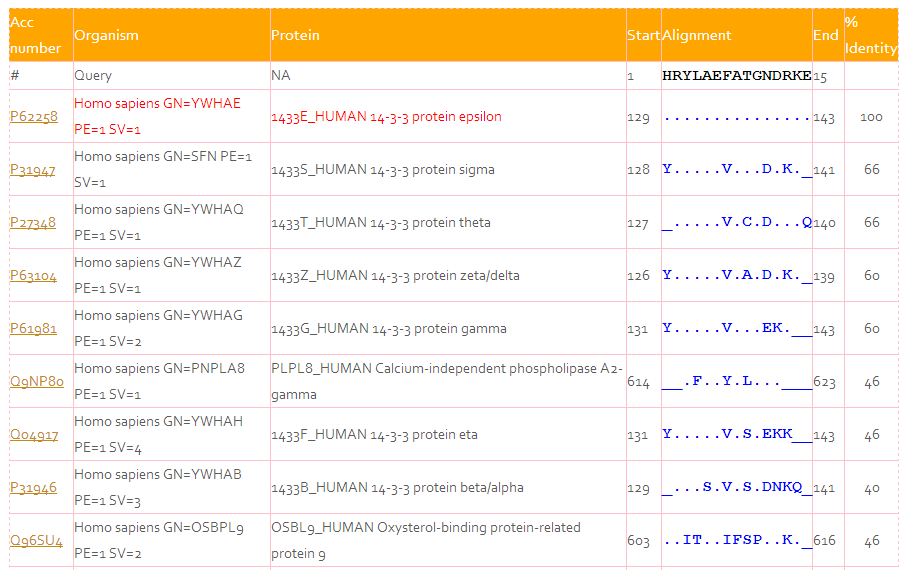
e) AVP-IC50 Conserve
Checks the conservation of user provided peptide sequence in human, viral and antiviral proteins