Prediction of peptide antiviral activity in terms of IC50

Algorithm
Machine learning Techniques:
Support vector machines (SVMs) were trained with the selected sequence features to predict peptide potency in regression mode. SVM allows choosing a number of parameters and kernels. The SVMlight software package (available at http://svmlight.joachims.org/) was used to construct SVM classifiers. In this study, we used the radial basis function (RBF) kernel:
k(x ,y)=exp(-γ||x - y||2)
where x and y are two data vectors, and γ is a training parameter.In addition we also used other machine learning methods like Random Forest (R package), IBk (Weka) and KStar (Weka) but the performance on SVM was best. Random forests are an ensemble learning method for classification (and regression) that operate by constructing a multitude of decision trees at training time and outputting the class that is the mode of the classes output by individual trees. IBk is a K-nearest neighbours classifier. It can select appropriate value of K based on cross-validation and distance weighting. KStar is an instance-based classifier, that is the class of a test instance is based upon the class of those training instances similar to it, as determined by some similarity function. It differs from other instance-based learners in that it uses an entropy-based distance function. Features:
a) Composition
Amino acid composition is the fraction of each amino acid in a peptide. The fraction of all 20 natural amino acids was calculated using the following equation:
Fraction of amino acid X=Total no. of X/peptide length
b) Database scanning
The sequence segments with high identity are inclined to share the structure and function. This method has been widely used in the past for proteins and peptide prediction, like in signal peptide and antimicrobial peptide prediction using BLASTP. We have also implemented BLASTP algorithm for prediction of AVPs. Each query sequence is matched against two newly released antiviral peptide databases viz. AVPdb and HIPdb described earlier.
c) Physicochemical properties
We tried all 544 physicochemical properties available in AAindex database individually, to look into the importance of each property in antiviral activity of the peptides. Finally selected 15 best performing physicochemical properties were used in developing AVPphysico model as provided below:
QIAN880113 , SUYM030101 , FINA910102 , CHOP780211 , MUNV940105 , TANS770102 , MUNV940104 , PALJ810114 , PALJ810115 , QIAN880110 , AURR980103 , ISOY800104 , MUNV940102 , RACS820110 , and AURR980105.
d) Binary profiles
Binary profiles were generated for the peptides with each amino acid being represented by a vector of 20 dimensions (e.g. Ala by 1,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0 and Cys by 0,1,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0 etc .) for the incorporation of positional information of amino acids in a peptide. A pattern of window length 'w' was represented by a vector of dimensions 20*w.
Evaluation:
In order to evaluate performance of our models, we used Pearson’s correlation coefficient (R). All models were evaluated using ten-fold cross validation technique.
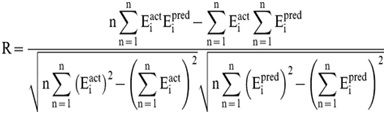
Where n is the size of test set, Eipred and Eiact is the predicted and actual IC50 respectively.