Database
VIRmiRNA is a resource for experimentally validated viral miRNA and its target reported in literature. It comprises of three sub-databases-VIRmiRNA, VIRmiRtar and AVIRmir. VIRmiRNA contains 1308 miRNA entries encoded by 44 viruses. While there are 7283 entries for VIRmiRtar and 542 entries for antiviral miRNA. The database also provides user-friendly search, advance search, browse options and also useful services like BLAST and Align .This viral miRNA resource has been designed for understanding host-viral interactions and development of antiviral therapeutics. This database is freely available through this url : http://crdd.osdd.net/servers/virmirna/.
Search
The search option is incorporated into all of the three sub-databases - VIRmiRNA, VIRmiRtar and AVIRmir. By selecting one option from three sub-databases, the user can enter the query in the box. This query search can be done in any of the given fields like virus name, miRNA name or miRNA sequence etc. or else user can select the “ALL” means query will be searched against all the fields.
Advanced Search
Here you can make more flexible queries by using ‘AND/OR’ to search the desired field. Likewise in search option , advance search is also available in all 3 separate sub-databases.
Browse
This links helps the user to explore the data in each sub-database catalogued/indexed for each virus like by clicking the VIRmiRNA under browse link.
Data Organization
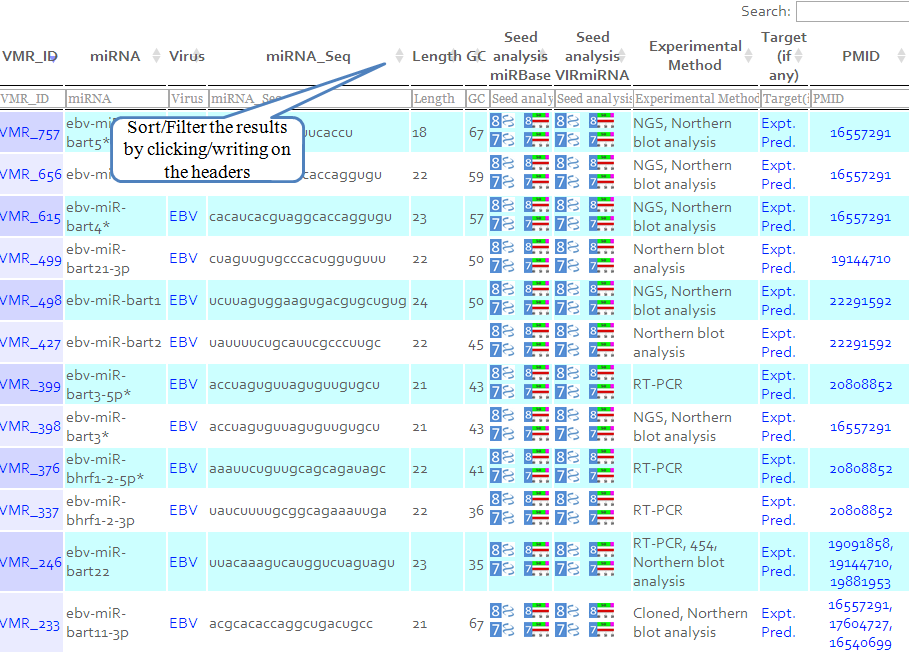
The data is organised as displayed below. After clicking on VIRmirID, you will be redirected to all the fields corresponding to this entry -Virus, miRNA name, miRNA sequence, Length, GC content, Family of virus, Target Gene.
Tools
i) BLAST
BLAST allows the alignment of user provided miRNA sequence against all the sequence available in our database. This help the user to confirm whether the given miRNA sequence have already been reported in the literature or not.
ii) Align
Here user can align given miRNA seed sequence with reference cellular/viral miRNA seed aequences.
iii) Map
Here user can map given miRNA with those in our database.
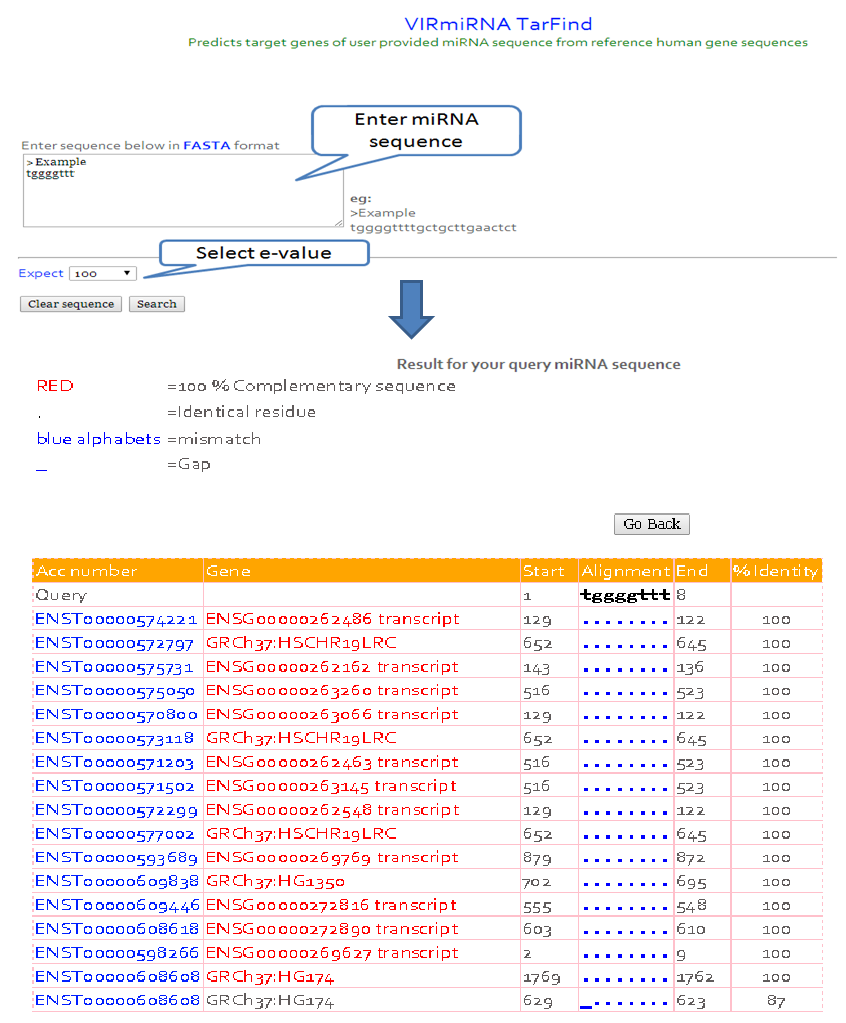
iv) Tar-Find
Predicts target genes of user provided miRNA seed sequence from reference human gene sequences.
v) Update
One of our main priority is to regularly update the database and addition of new viruses.