Datasets-
The datasets were derived mainly from Hemolytik database. We have developed two following datasets:
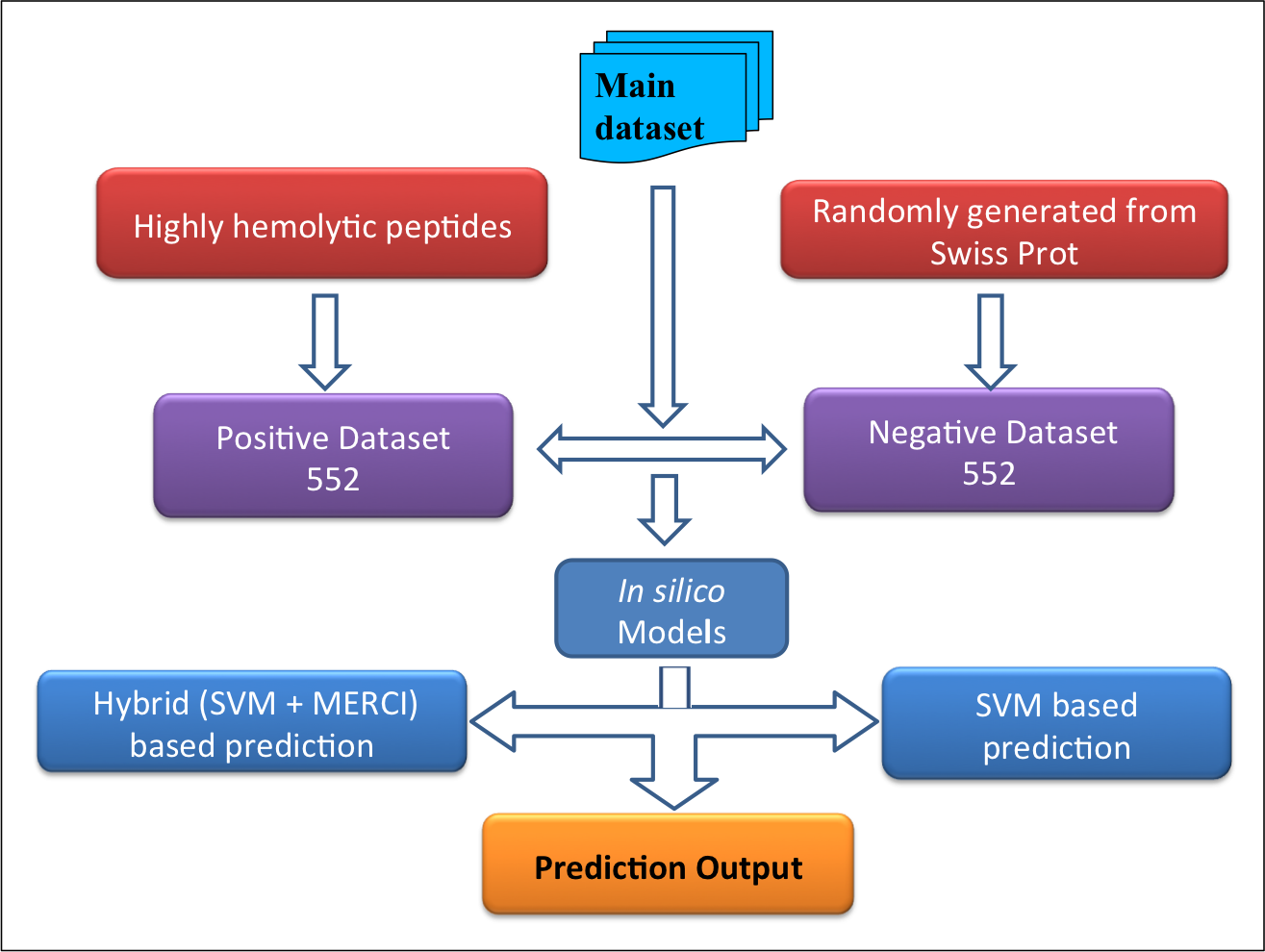
Main dataset-
This comprises of 552 experimentally validated highly hemolytic peptide sequences from Hemolytik Database as positive dataset and same number of peptide sequences randomly generated fron SwissProt as negative dataset as shown in following figure. |
|
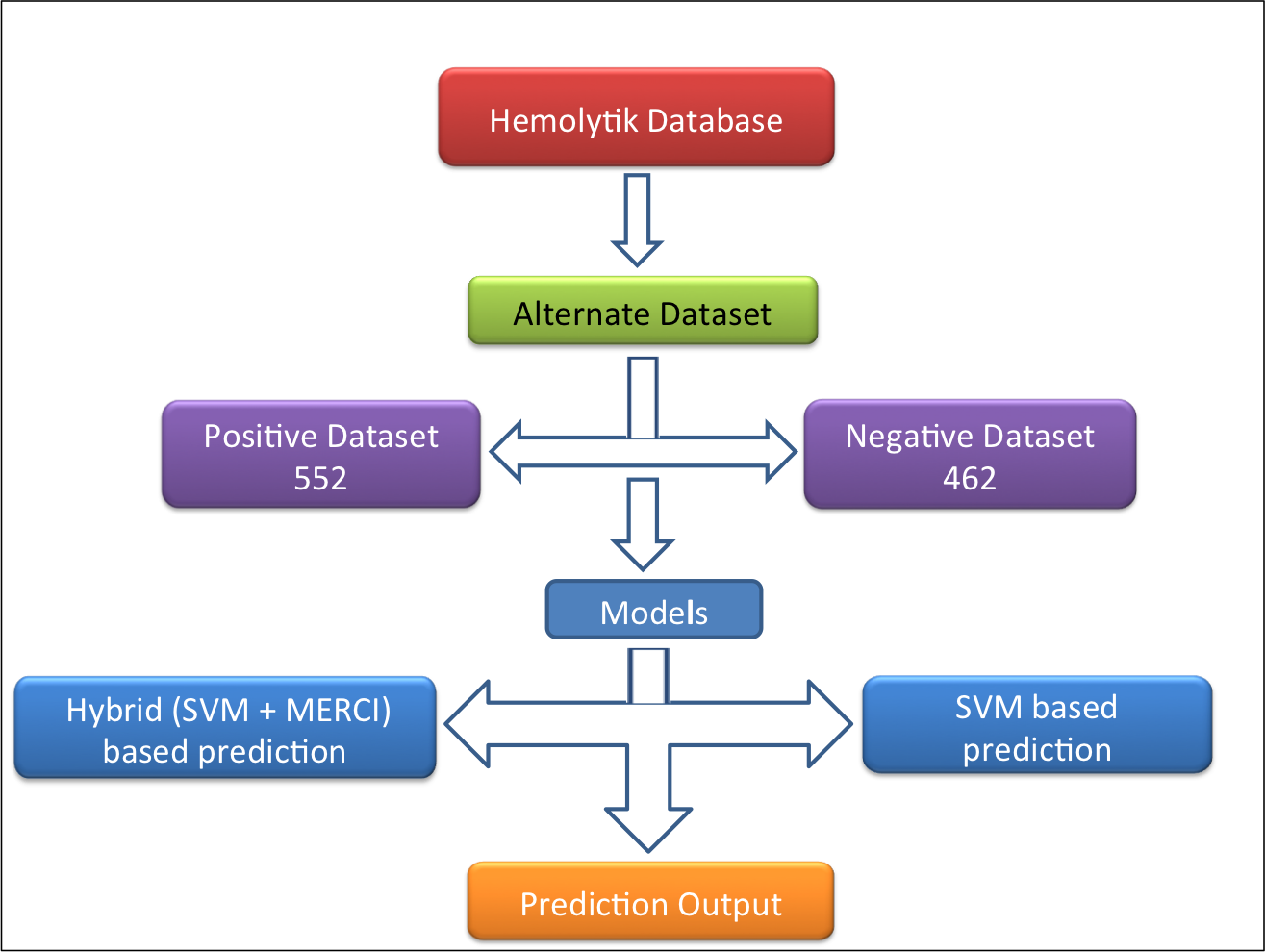
Alternative dataset-
In this dataset (alternate dataset), 552 experimentally validated highly hemolytic peptides were taken as positive examples and 462 experimentally validated non-hemolytic/poor hemolytic peptides (i.e. peptides having IC50>200uM, <10% hemolysis at 100 or >100uM concentration and MHC >500uM) were taken as negative example as shown in following figure.
|
|
|
The algorithm of HemoPI is based on the following models: |
Motif based model |
| A number of motifs were observed in hemolytic peptides. Therefore, we used this information for differentiating between hemolytic peptides and non-hemolytic peptides.
These motifs were used for distinguishing hemolytic peptides from non-hemolytic peptides. |
Hybrid approach (Motif and SVM)
To increase the robustness of prediction, we used the hybrid model by integrating motif based and SVM based methods. If the user's query peptide has the any one of the motifs as those from our datasets, we assign peptide type to it based on motif that it contains. SVM model of prediction is used in case no motif is found.
|



