Help Page of PEP2D
This page provides help on each web page of PEP2D. On following menus/submenus help is provided with screen shorts.
Prediction Module |
Input sequence to Prediction Module
User can enter peptide sequences in fasta format in the input box. The server also accepts multiple sequences in fasta format (max. 10 sequences at a time).
User can also upload a file having multiple sequences in fasta format (max. 10 sequences at a time).
If email address is provided, the results will be mailed to the provided email address.
If you have multiple peptides (more than 10 sequences) you can use our "Multiple Sequences" Module for prediction of secondary structure of multiple peptides. (In this module there is a limitation of 1000 sequences which can be given as input to the server).
The Figure below represents how to input peptide sequences in the prediction module of PEP2D server as well as the display of results in text and graphical format.
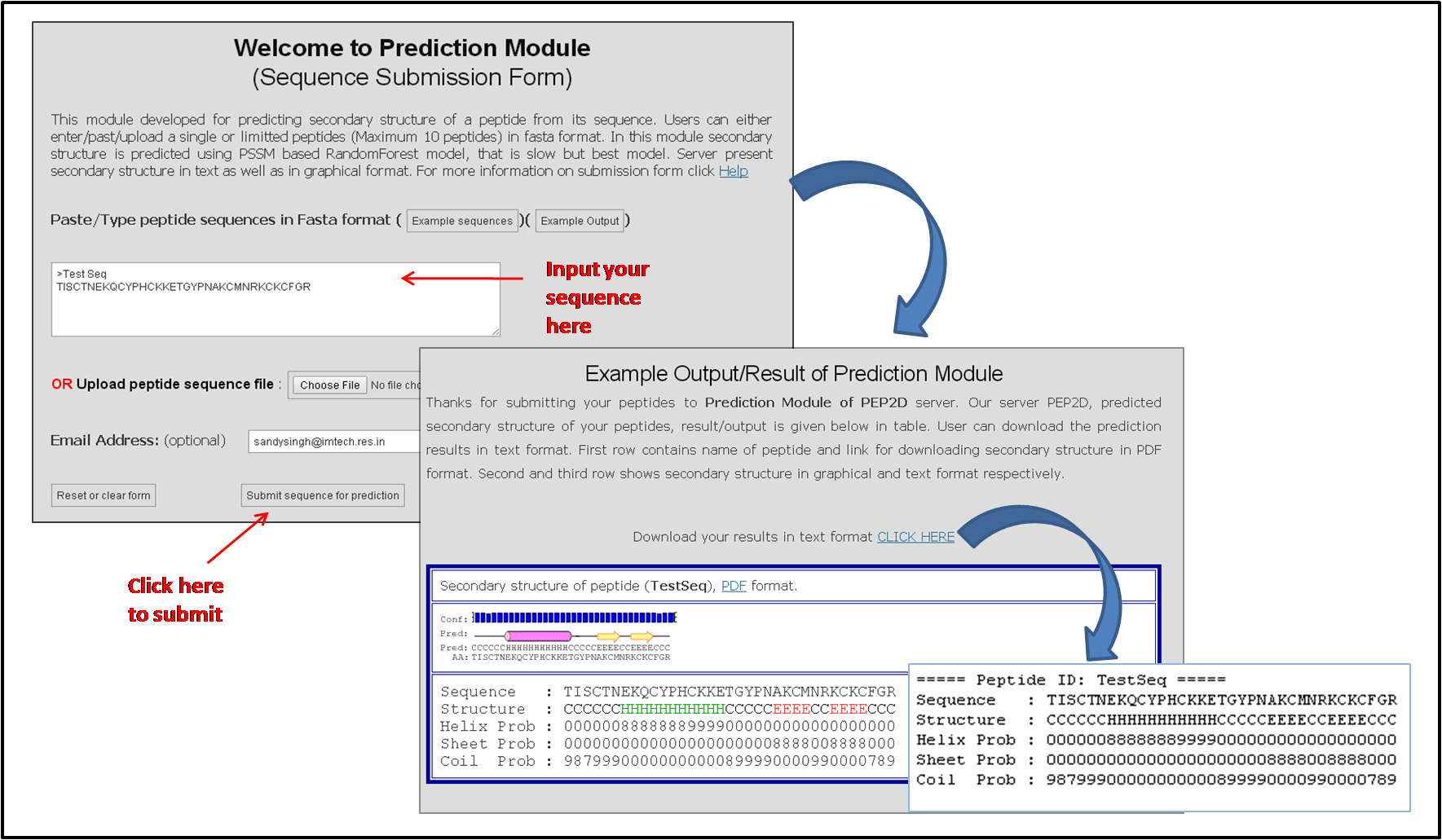
Figure 1: PEP2D prediction module representing how to input a peptide sequence for its secondary structure prediction.
Results of Prediction Module
The results of prediction module are provided in both text as well as graphical representation as shown in the above Figure 1. A link is provided to download all the results in the text format. In graphical representation, helix is represented in cylindrical shape in pink color and sheet is represented as an arrow in yellow color where as coil is represented as black line. Confidence values are represented as bars where increasing length of bar represents increasing confidence level. This graphical representation is also represented in PDF format.
Multiple Sequences Module |
Input sequence to Multiple Sequences Module
User can enter peptide sequences in fasta format in the input box. The server also accepts multiple sequences in fasta format (1000 sequences at a time).
User can also upload a file having multiple sequences in fasta format.
If email address is provided, the results will be mailed to the provided email address.
The Figure below represents how to input peptide sequences in the multiple sequences module of PEP2D server.
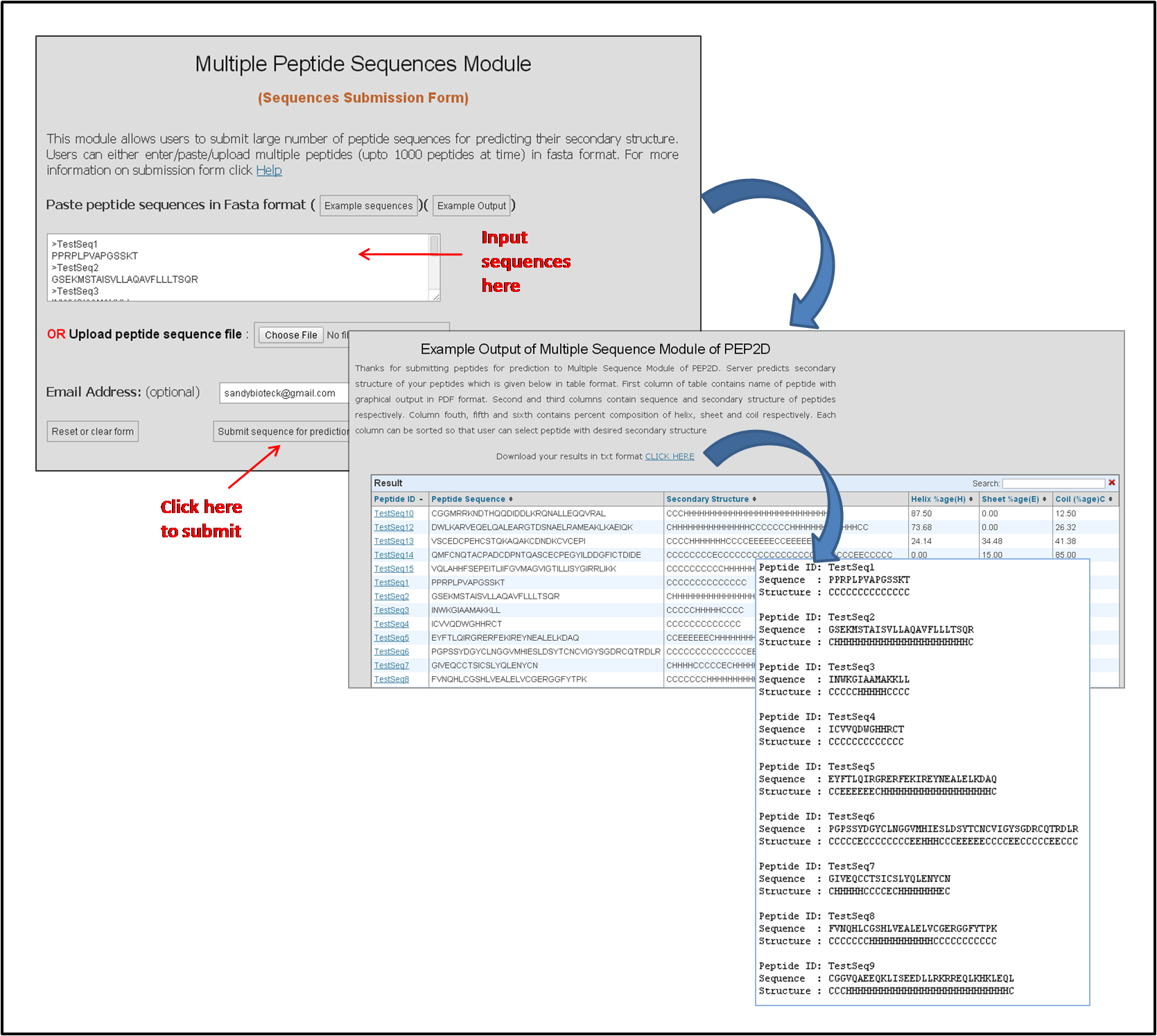
Figure 2: PEP2D multiple sequences module representing how to input a peptide sequence for the secondary structure prediction of multiple sequences and the representation of results of this module in text and graphical format.
Results of Multiple sequences Module
The results of multiple sequences module are provided in both text as well as tabular representation as shown in the above Figure 2. A link is provided to download all the results in the text format. In tabular representation, first column contains peptide ID which is hyperlinked with the PDF file containing graphical representation of the results. Placing mouse over the peptide id shows an image with the same graphical representation as given in the PDF file. Second Column contains peptide sequence, third column represents predicted secondary structure of peptides, 4th, 5th and 6th column represents percent composition of helix, sheet and coil in that peptide.
Mutant Peptides Module |
Input sequence to Mutant Peptides Module
User can enter peptide sequences in plain text format in the input box. This module generates all the mutant peptides at each position of the input peptide sequence and predicts the secondary structure of each mutant. In this way a user can select peptides with desired secondary structure.
If email address is provided, the results will be mailed to the provided email address.
The Figure below represents how to input peptide sequence in the mutant peptides module of PEP2D server.
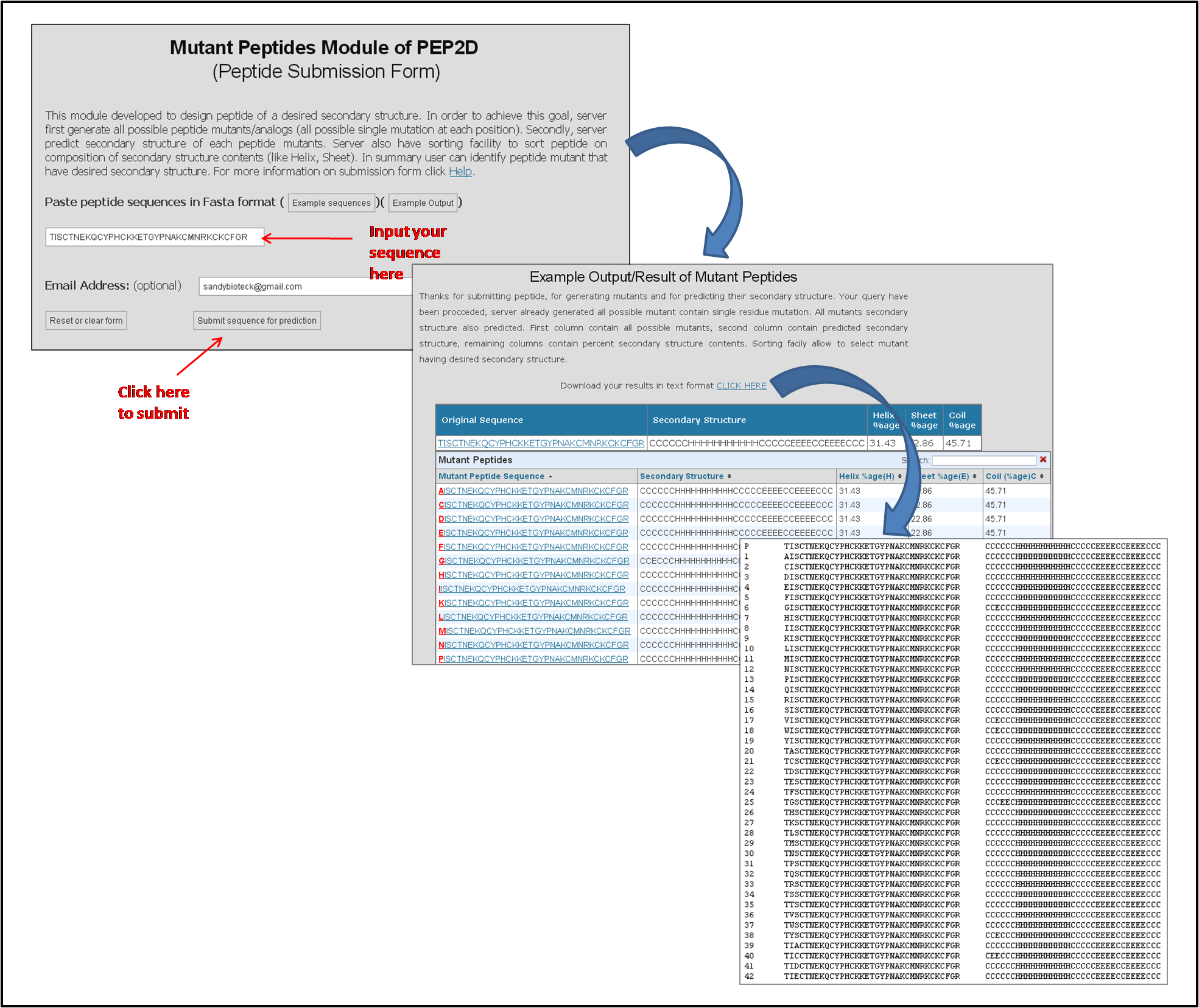
Figure 3: PEP2D mutant peptides module representing how to input a peptide sequence for the secondary structure prediction and its result representation in text and graphical format.
Results of Mutant Peptides Module
The results of mutant peptides module are provided in both text as well as tabular representation as shown in the above Figure 3. A link is provided to download all the results in the text format. In tabular representation, first column contains peptide ID which is hyperlinked with the PDF file containing graphical representation of the results. Placing mouse over the peptide id shows an image with the same graphical representation as given in the PDF file. Second Column contains peptide sequence, third column represents predicted secondary structure of peptides, 4th, 5th and 6th column represents percent composition of helix, sheet and coil in that peptide. Each position which is mutated is represented by red color.