
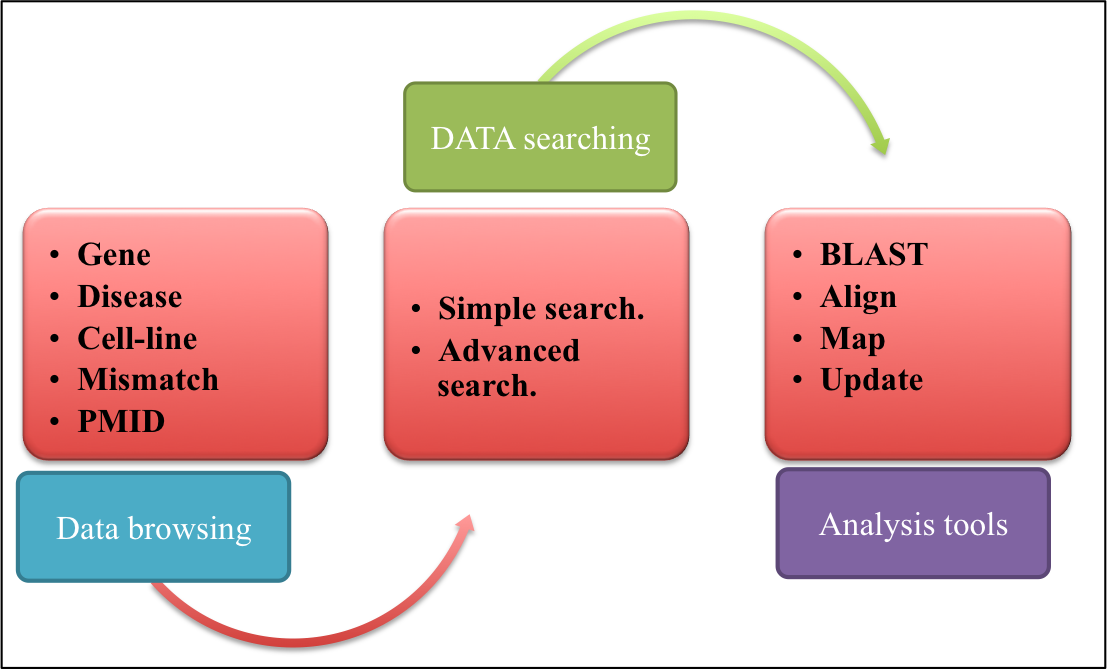
CONTENTSDatabaseBrowse Search Advanced Search Data organization Tools Update |
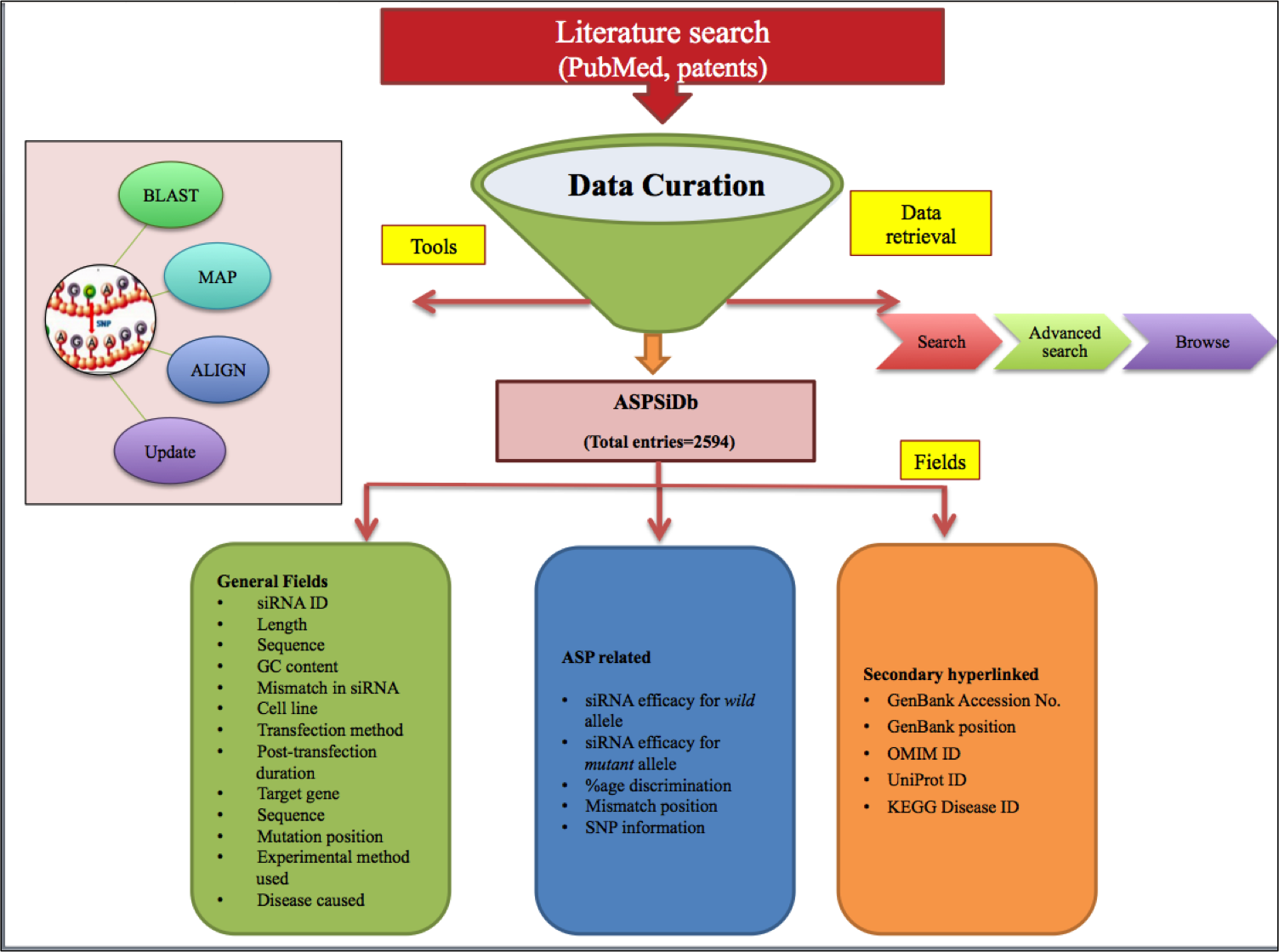
ASPsiDb is a comprehensive repository of experimentally validated Allele specific siRNAs (ASP-siRNAs) targeted against 62 different genes from 43 different human disease. As name indicates, ASP-siRNAs; unlike allele indiscriminative general siRNA, they are capable of differentiating the two alleles/forms of a target gene even if it differ only with one single nucleotide difference. Thus they can be harnessed to use in the suppression of gene expression with mutant form, in such a fine tuned way that wild type counterpart of gene will be spared or minimally harmed. Presently, this repository contains experimentally verified 2594 ASP-siRNAs.ASPsiDb contains ASP-siRNAs coming from varied percentage discrimination and efficacies and tested on 30 different cell lines.Database also provides some informative tools for data analysis such as BLAST, Align and MAP as well as links to major siRNA resources. Not just a archive of ASP-siRNA sequences, but database is designed with the aim to increase our understanding of Human Diseases by mutations and provide maximum useful information to the user; via different tools and external links like OMIM ID, KEGG disease ID, UniPROT ID, Ensembl ID. This database is freely available through this url http://crdd.osdd.net/servers/aspsidb. The database comes with enhanced user experience and easy to operate browsing as well as searching with sorting and filtering functionalities. This database has been dedicatedly designed, referring to the current needs of scientific community working exhaustively for RNAi therapeutics development and is anticipated to be useful for them who are seeking a committed repository for ASP-siRNAs.
.
User can perform search option in the following two ways:
2) Second
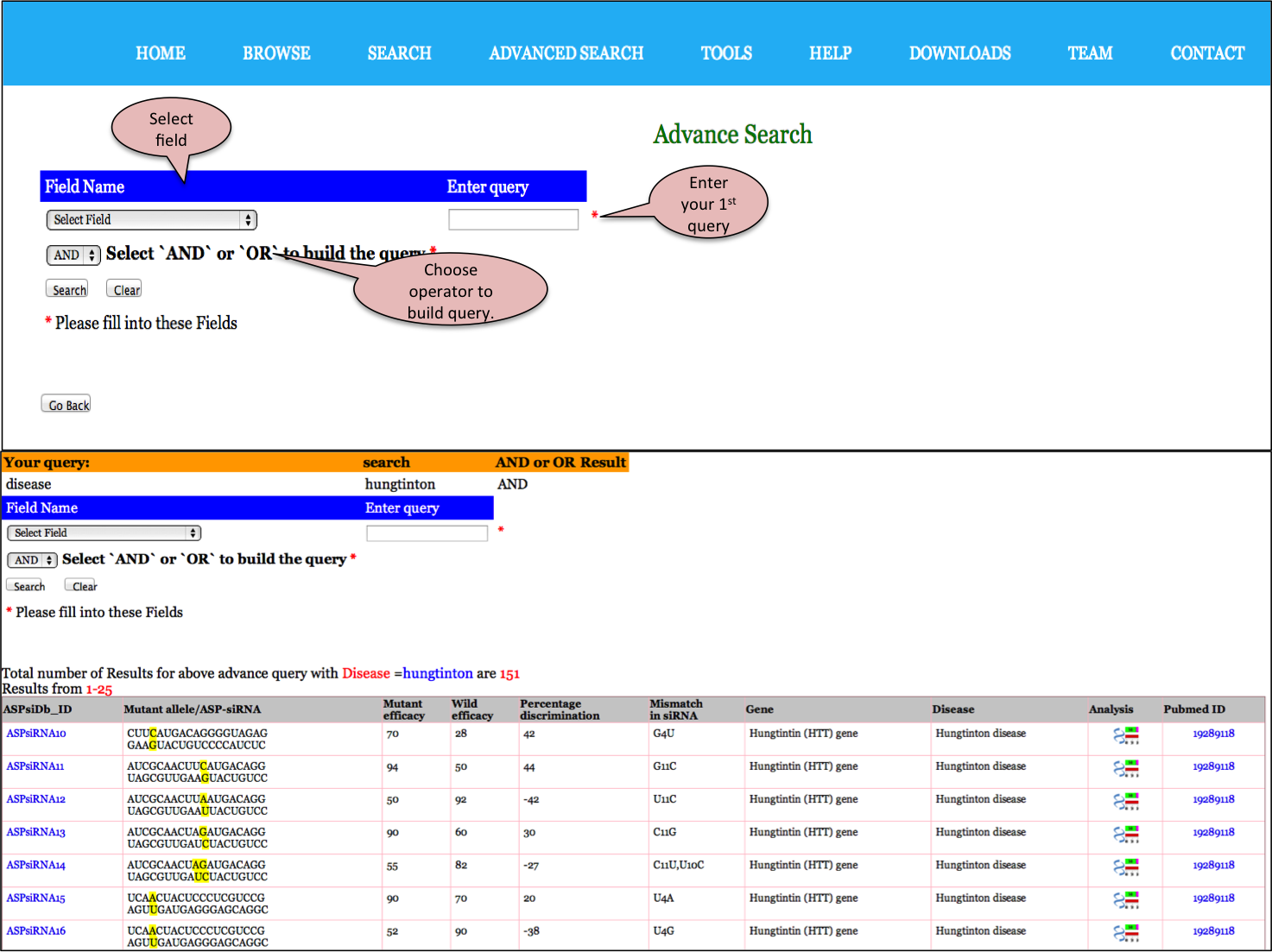
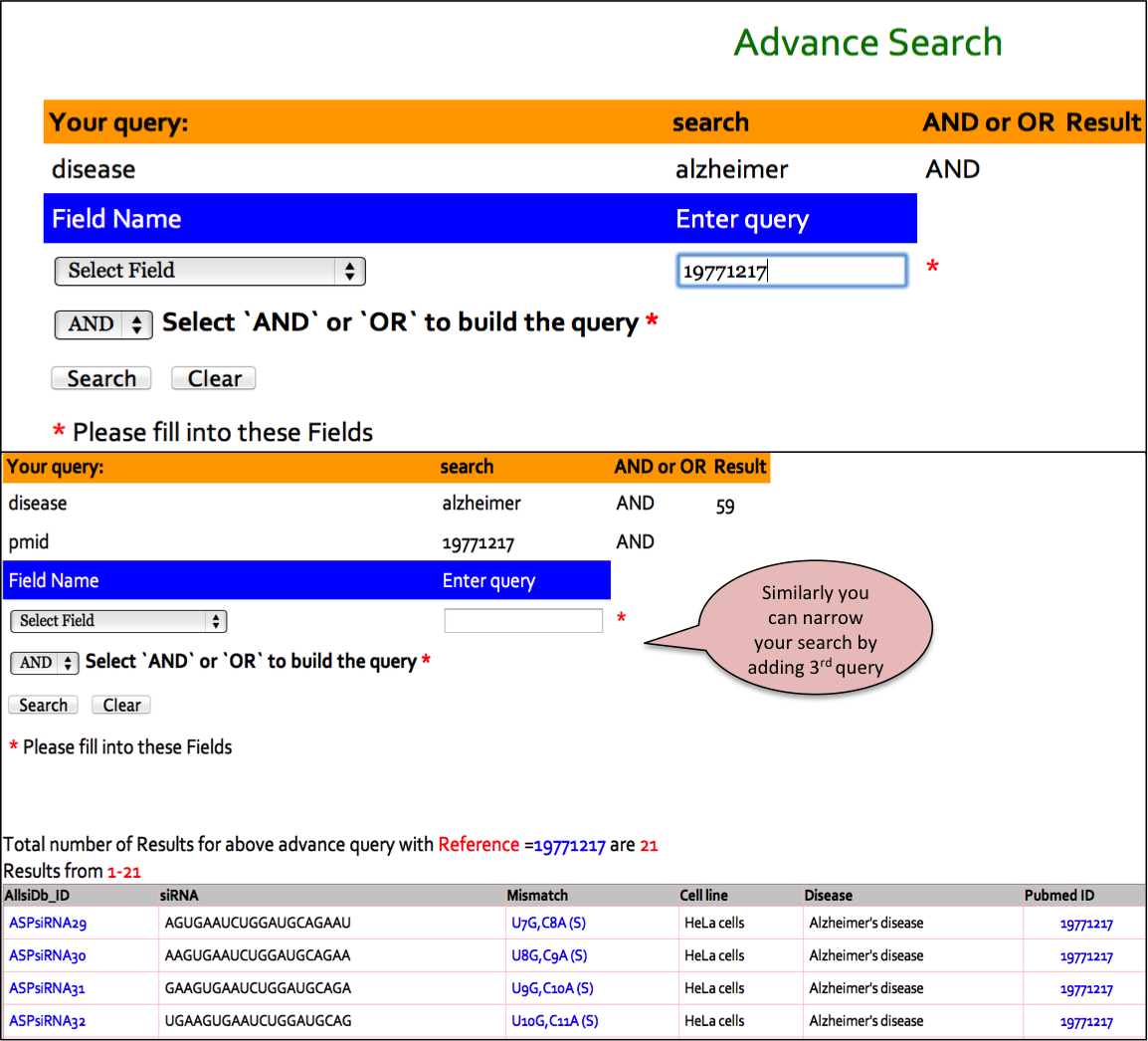
Input: This feature helps you to search this database using logical operators (AND/OR). For example, you have obtained say 151 results for ASP-siRNAs targeted against Hungtintin gene. If you wish to further filter out those ASP-siRNAs with PubMed ID as <19412185.In that case,you can use AND operator alongwith choosing the Reference as desired field.
Output: In the example below results obtained from this search will retrieve the AVPs corresponding to Virus abbreviation = WNV and Assay = Plaque assay and Peptide length = 19.
user can enter the query/keyword after clicking search on menu bar.The user given keyword can be searched in the all fields like siRNA ID,SiRNA sequence sense,Mismatch in siRNA,Disease,Concentration,Target,Mismatch in Target, UniProt ID, Target sequence, SNP,%age discrimination, Cell line, Experimental technique, siRNA expression method, OMIMID, PMID, Transfection method OR there is also facility that user can select some out of all fields to search that keyword in those fields only.
1) User can perform a simple search by giving a keyword to the radio button provided on the seacrh page and click submit.
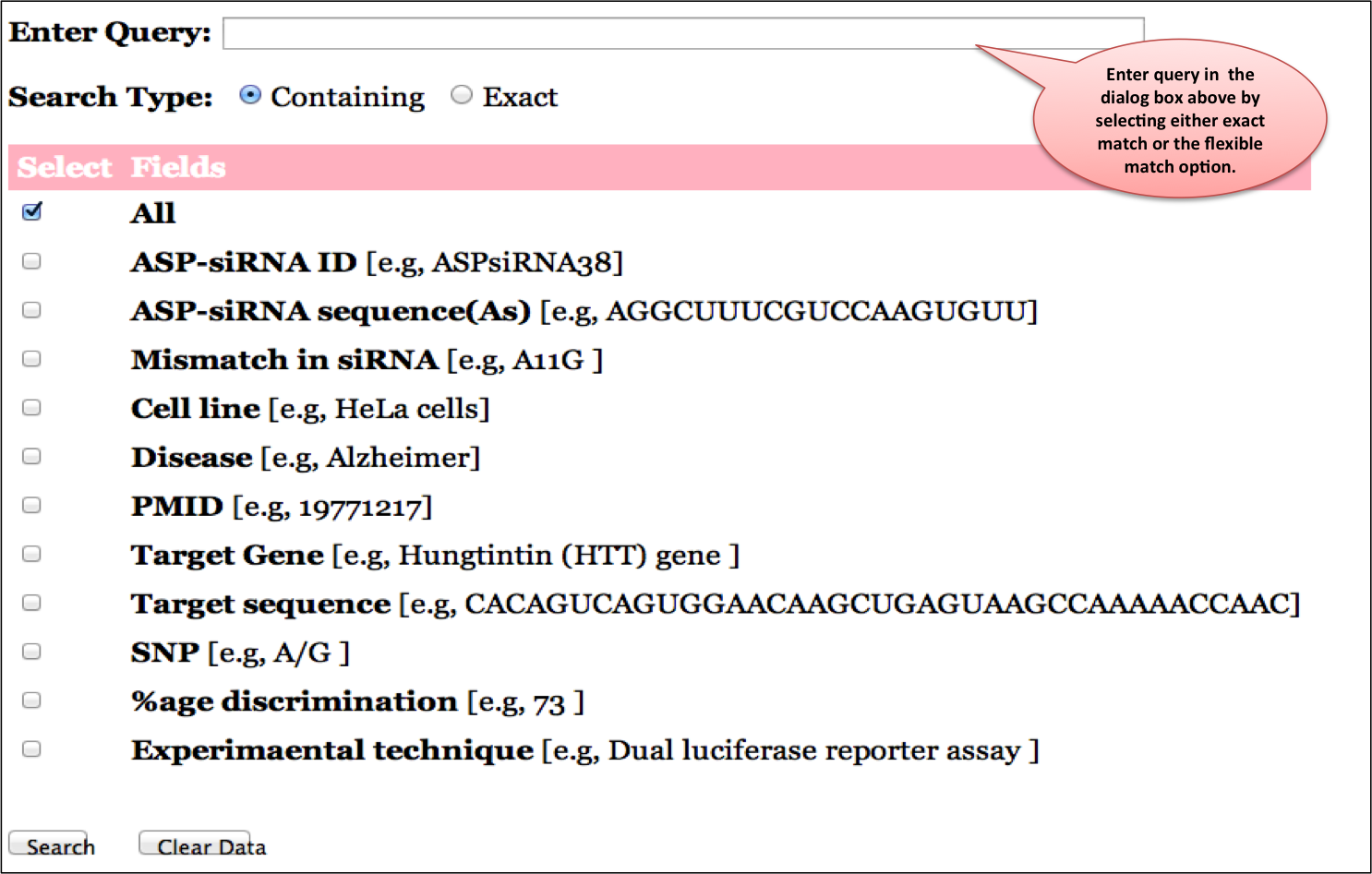
Input: Here you can enter your query in the box and can specify any of the 11 fields like ASP-siRNA ID,ASP-siRNA sequence (Antisense),Mismatch in siRNA,Disease,PMID,Target,Target Gene,Target sequence, SNP,%age discrimination,Experimental technique against which you wish to search or else you can keep the default All option which will search against all the fields in the database. Examples have been provided corresponding to each field which can be used as sample queries. Besides the option to choose the field, search type allows to retrieve either an exact match or the match containing the query. Clicking on clear data button clears the data after your search.
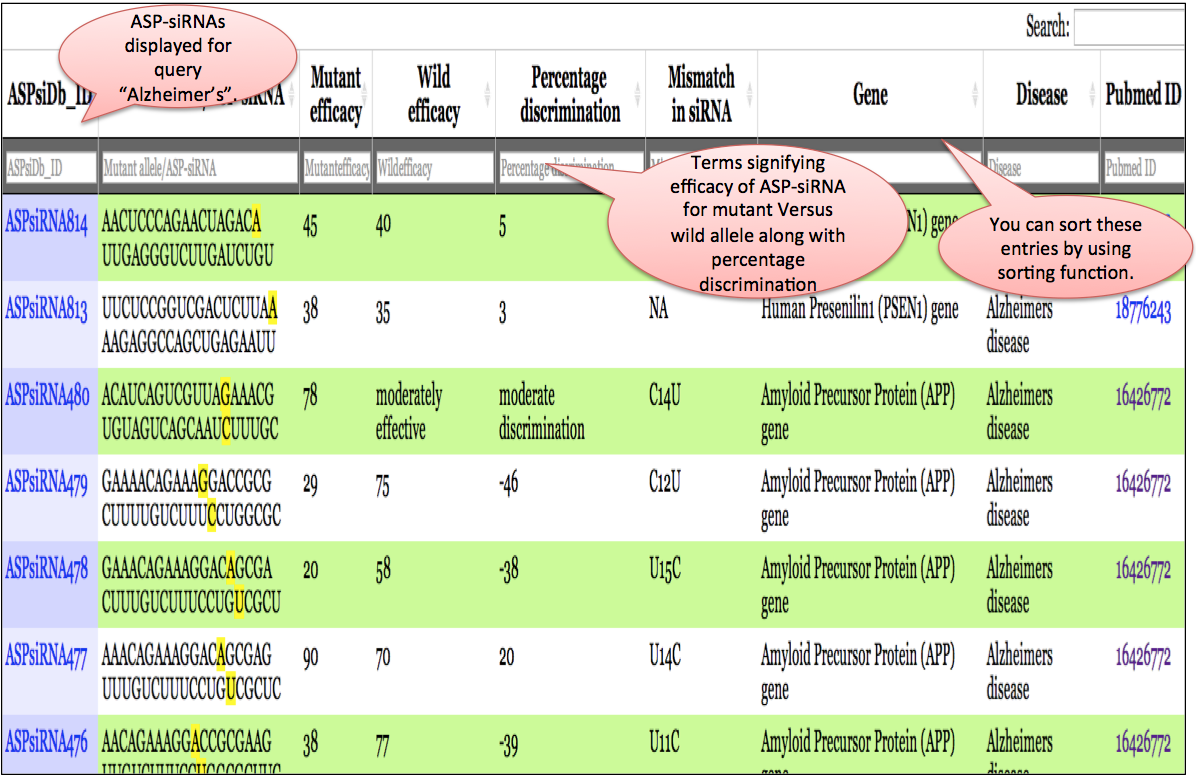
Output: The results obtained from this search display 10 fields where 9 fields contain the experimental data and the one, Analysis, has links to BLAST and align results. The entries in blue colour are further hyperlinked like ASPsiRNA814 is linked to further experimental details of this ASP-siRNA, wgile links to gene, diseaseand PMID redirects you to UniProt, OMIM and PubMed respectvely. You can sort the data by clicking on the column header and filtering can be done by typing in the boxes given under the headers.
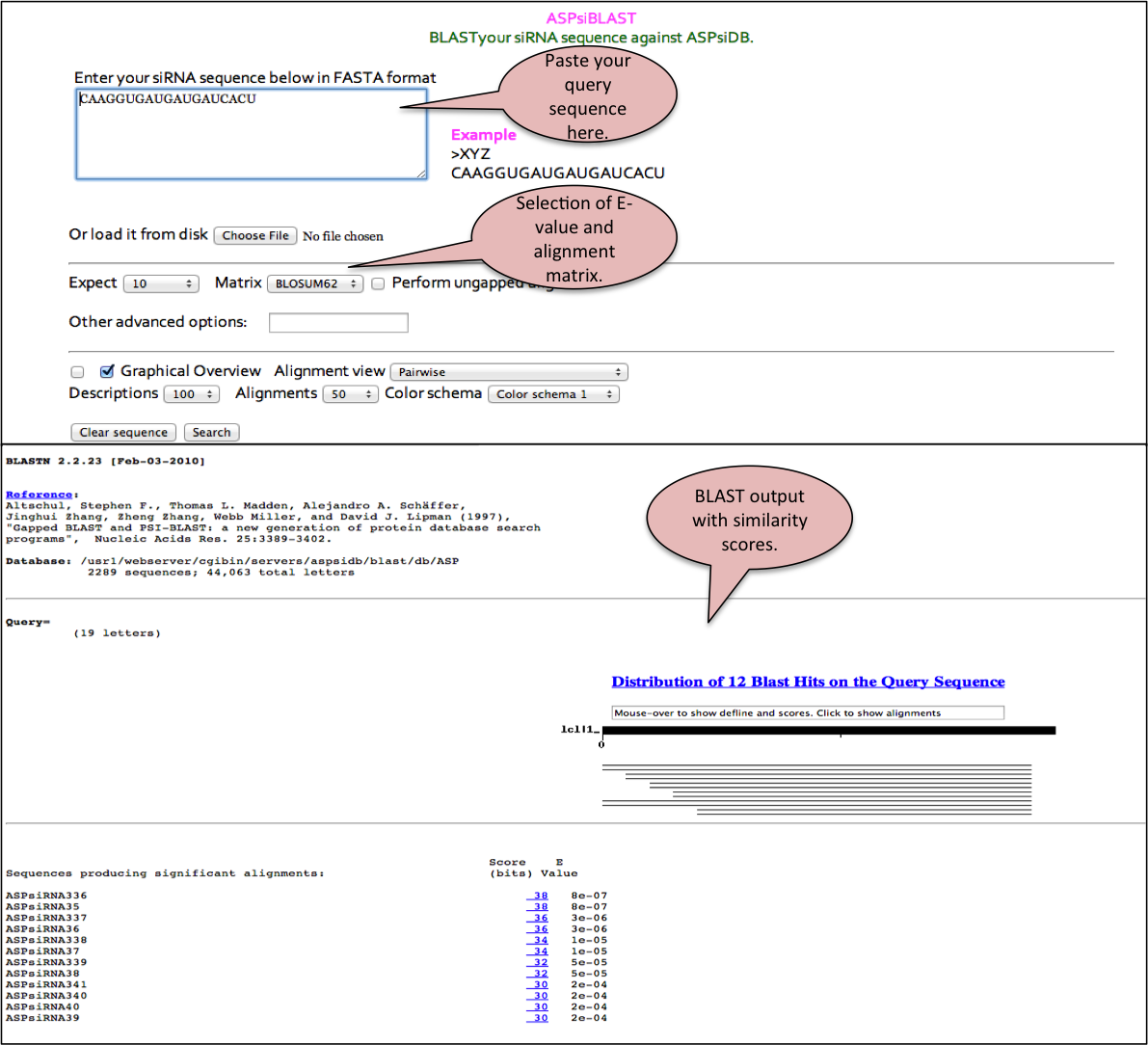
ASPsidb Blast
Additionally, the Blast allows alignment of a user provided siRNA sequence against all the peptide sequences available in our database. This helps the user to confirm whether a given peptide sequence or similar one has already been reported or not.
The output is given in the standard format with the blast score and e-value. The alignment is shown for the peptides found to be identical or similar in the database. The output can be formatted based on the options provided by user.
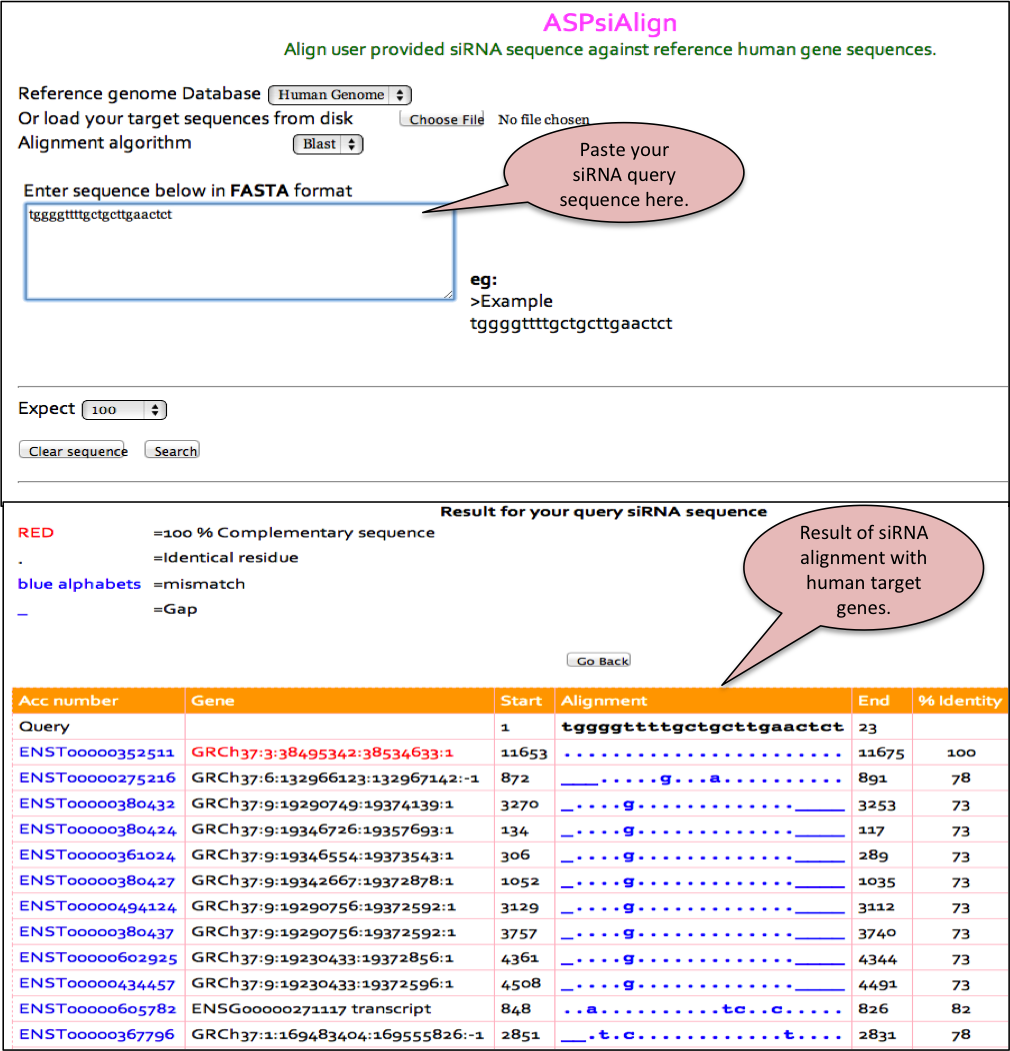
ASPsidb Align
Additionally, the Blast allows alignment of a user provided peptide sequence against all the peptide sequences available in our database. This helps the user to confirm whether a given peptide sequence or similar one has already been reported or not.
The output is given in the standard format with the blast score and e-value. The alignment is shown for the peptides found to be identical or similar in the database. The output can be formatted based on the options provided by user.
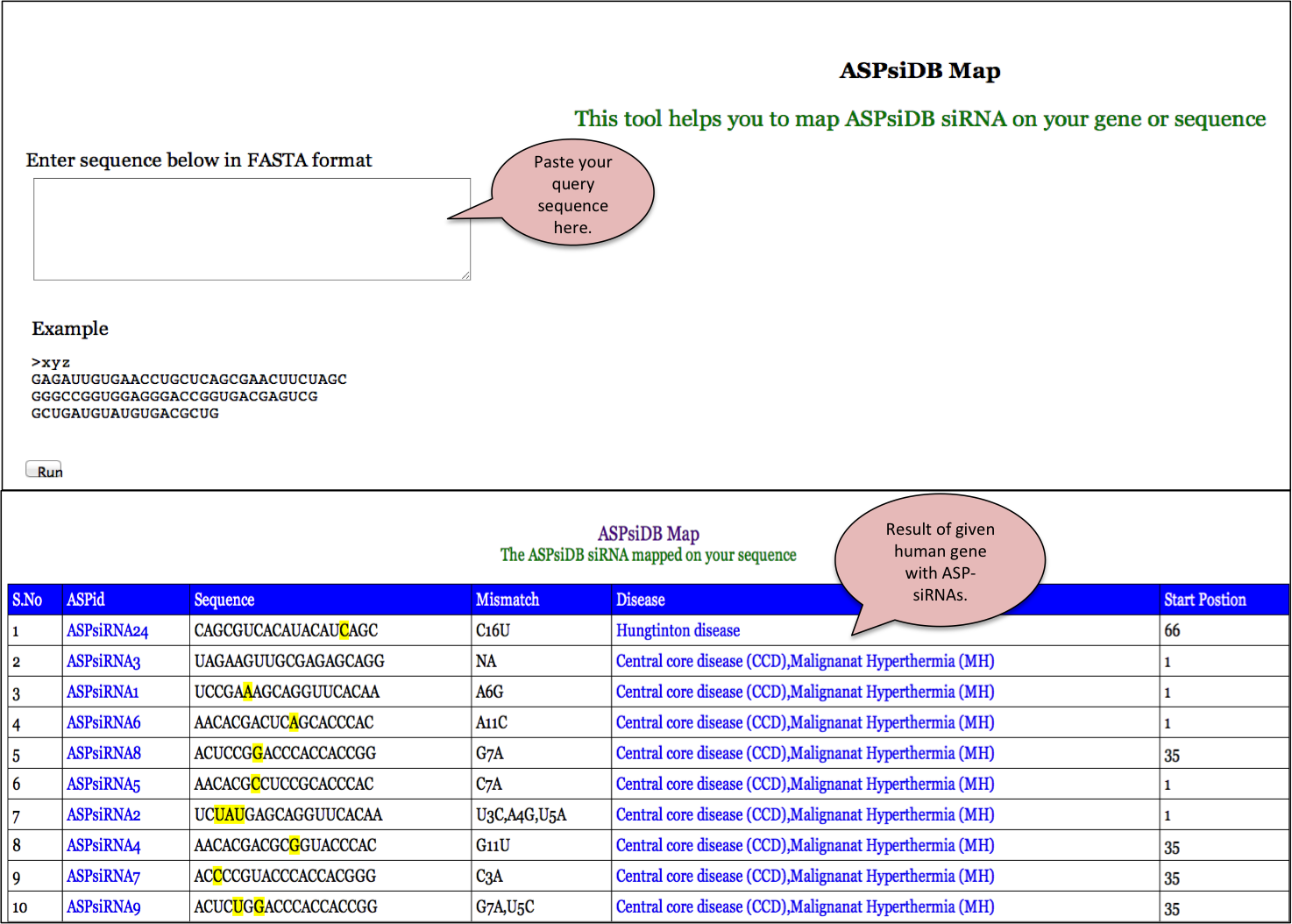
ASPsidb Map
Additionally, the Blast allows alignment of a user provided peptide sequence against all the peptide sequences available in our database. This helps the user to confirm whether a given peptide sequence or similar one has already been reported or not.
The output is given in the standard format with the blast score and e-value. The alignment is shown for the peptides found to be identical or similar in the database. The output can be formatted based on the options provided by user.
Authors generating experimental data on ASP-siRNAS are encouraged to submit the data directly into ASPsiDb. For this purpose, a web form for data submission is provided. Submitted information will be included in the database update after ascertaining its authenticity.