AVPpred INPUT
Paste your single or multiple peptide sequence(s) in FASTA format in the provided text-box or upload a FASTA file from your system. User can remove the entries in the text-box by using the clear button. An “Example link” has been provided to help the users. By clicking on Example link peptide fasta sequences will be automatically filled in the text-box. User can click the “Predict AVP” button to make antiviral peptide predictions. Default machine learning technique is SVM, however user have the options to run ANN and Random Forest also. Peptide features used for making AVPpred models are also shown.

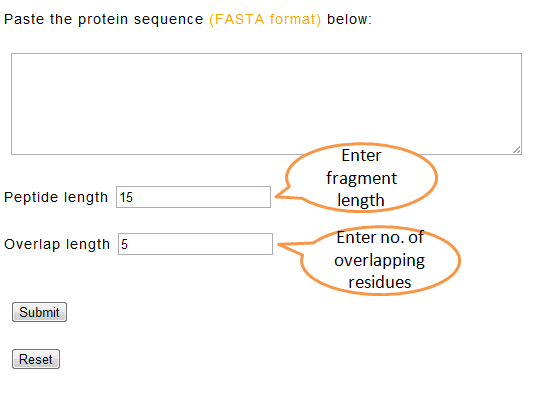
If user has a protein sequence, then, an option for making peptide fragments of desired length and overlapping residues is provided. User can paste these peptide fragments in the “Predict AVP” form. Note that you can enter only one protein sequence at a time. The fragments of desired length may then be used in the submit form at one go.
AVPpred RESULT OUTPUT
Results of the prediction will be shown in a tabular as well as graphical layout.
In table 7 columns are given. In first three columns Sequence No, Peptide Sequence Name and Length are given. Hovering on the sequence name (fasta header) in the table displays its sequence.
In fourth column BLAST option is provided to search similar peptide sequences in CAMP and AMP antimicrobial peptide databases and also in SWISS-PROT.
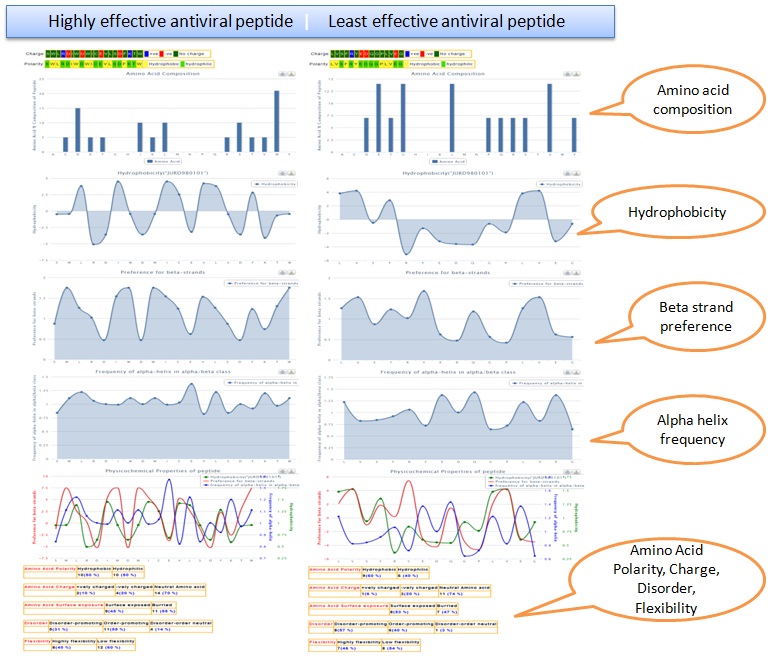
By clicking the next column, some relevant properties of the peptide like charge, polarity, amino acid composition, hydrophobicity, preference for alpha/beta strand, etc will be displayed.
Last columns of AVPpred prediction output i.e. Motifs, Align, Compo and Physico are based on antiviral motifs, Sequence alignment, Amino acid composition, Physicochemical properties models. In each cell of these column displays the numerical value of SVM score. 100 is the best and 0 is the least. This is displayed in color code from red to green. Sorting option is provided to help the users in sorting the data result output. Simultaneously a search option is also provided to filter the data.
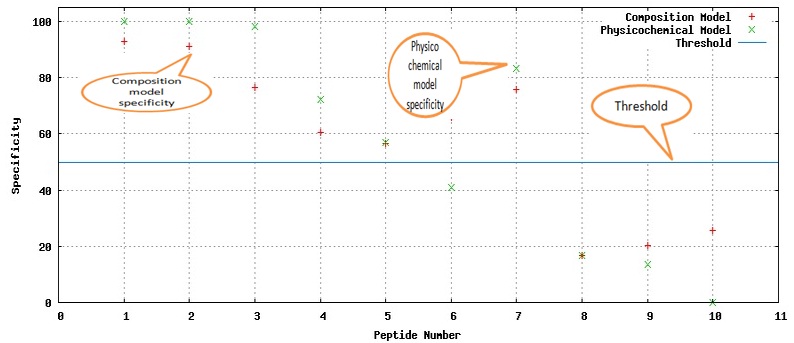
The Graphical View of the Result displays the specificity of the peptides corresponding to the Composition and Physicochemical models. Blue line shows the default threshold of 50. Note that this graph is shown for multi-fasta files.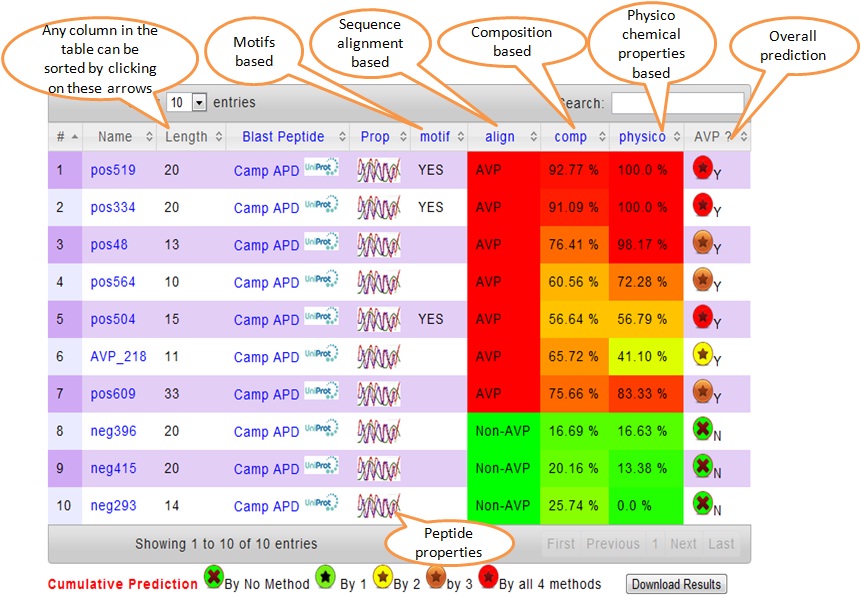 Graphical View of the Result
Graphical View of the Result
Result output of BLAST showing distribution of hits, their score, e-value and alignment with sequences with significant similarity will be displayed. By clicking the next column, some physico-chemical properties will be displayed.



ANALYSIS TOOL
Also the user can blast his own peptide sequence against the selected peptide databases through the blast form. Also user can check the physicochemical properties or predict peptides from other databases (CAMP and APD2)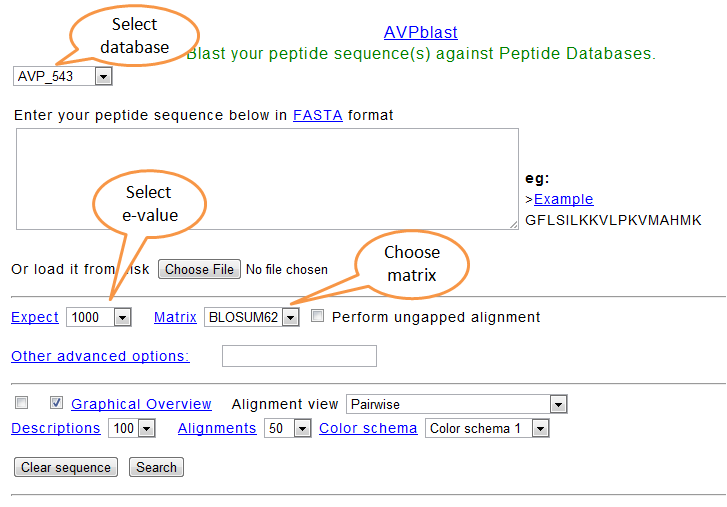
Further by clicking on "Properies" in side menu, some relevant properties of the user provided peptide like charge, polarity, amino acid composition, hydrophobicity, preference for alpha/beta strand, etc will be displayed.
Comparison of physico-chemical properties of most effective
antiviral peptide and the least effective one.