Tetrapeptide: Protein Scan
Protein Scan generates peptides using a 'sliding window' of fixed length from a query protein sequence. The fragments thus generated are subjected to prediction whether these would be antihypertensive or not.

|
All in One: Variable length I/P
The models used for prediction of antihypertensive peptides in this tool have been trained on peptides of all the lengths found in literature. In other words, this prediction tool uses a model that is applicable for query peptides of any length.

|
|
Propensity Matrices: P.Matrices
The tendancy of a specific amino acid to occur at a specific position in the peptide can be represented on a propensity scale. The Propensity Matrix displayed by this tool has the peptide positions as columns and amino acid as rows in the tabular form where each value represents the propensity of the amino acid to ccur at that position in the peptide.

|
|
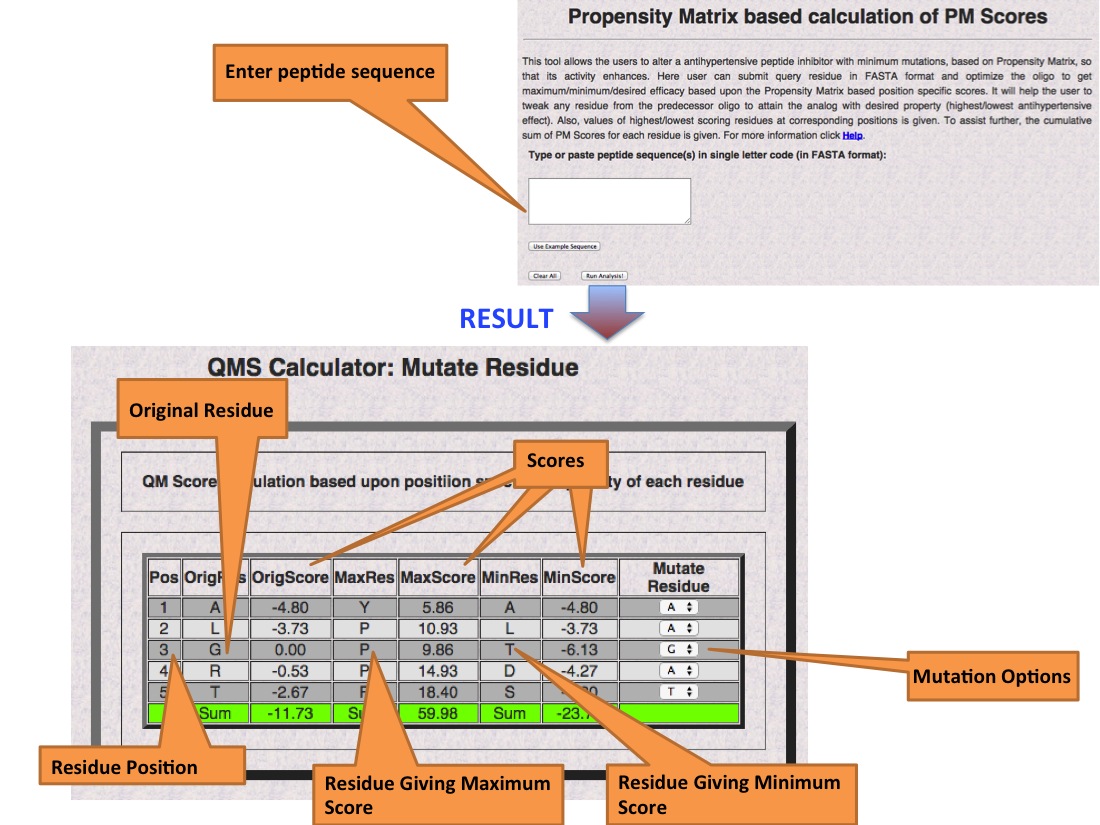
Propensity Matrices: PM Score Calculator
The PM Score Calculator allows the user to dynamically mutate residue(s) to try different combinations of amino acids in the sequence and observe the change in score as compared to the score of the original sequence. This allows the user to tailor changes that may increase or decrease the antihypertensive activity of the peptide.
|

