|
Home
Algorithm
Datasets
References
Developers
Contact
Important Sites
|
Algorithm behind Domprint
|
The Domprint SVM model has been trained on highly curated dataset of 5227 known DDI (positive dataset) and 4089 non-interacting domain pairs (negative dataset). The positive dataset is taken from 3did database (Stein et. al. 2005). The negative dataset has been compiled from 3did and PDB databases. The following figure illustrates the scheme adopted for making dataset-
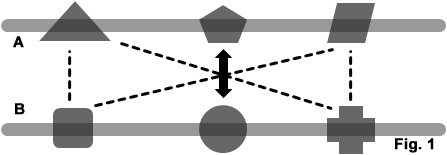
Figure 1: Schematic representation of dataset creation. Let A and B are two polypeptide chains in a protein structure, mapped with corresponding domain families. Biheaded arrow shows interaction between two domains taken from 3did database. Other domain pairs from A and B (linked by dashed lines), not knwon to be interacting in 3did database, constitute negative dataset used in this study.
|
|
Support Vector Machine (SVM) needs a fixed length pattern for training and testing of data. We calculated the amino acid composition for each domain in the dataset, and concatenated the composition frequency of domains of a pair.
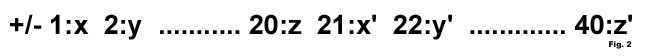
Figure 2: The feature vector for a domain pair (interacting or non-interacting). Plus sign for interacting pairs and vice-versa. Feature length is 40, 20 from each domain.
|
|


