

Allele-specific siRNAs (ASP-siRNAs); unlike general siRNAs, are capable of differentiating the between mutant/variant and wild-type allele of a target gene even if there is only one single nucleotide difference. Thus they can be harnessed to use in the suppression of gene expression of toxic, dominant, gain-of-function mutations/variants, in such a fine tuned way that wild-type counterpart of gene will be minimally harmed. ASP-RNAi approaches are emerging as feasible therapeutics that offer an alternative technique to modulate traditionally 'undrugable' targets especially in the case of dominantly inherited GOF neurodegenerative diseases, ASP-siRNAs can target the underlying cause of these intractable disorders.
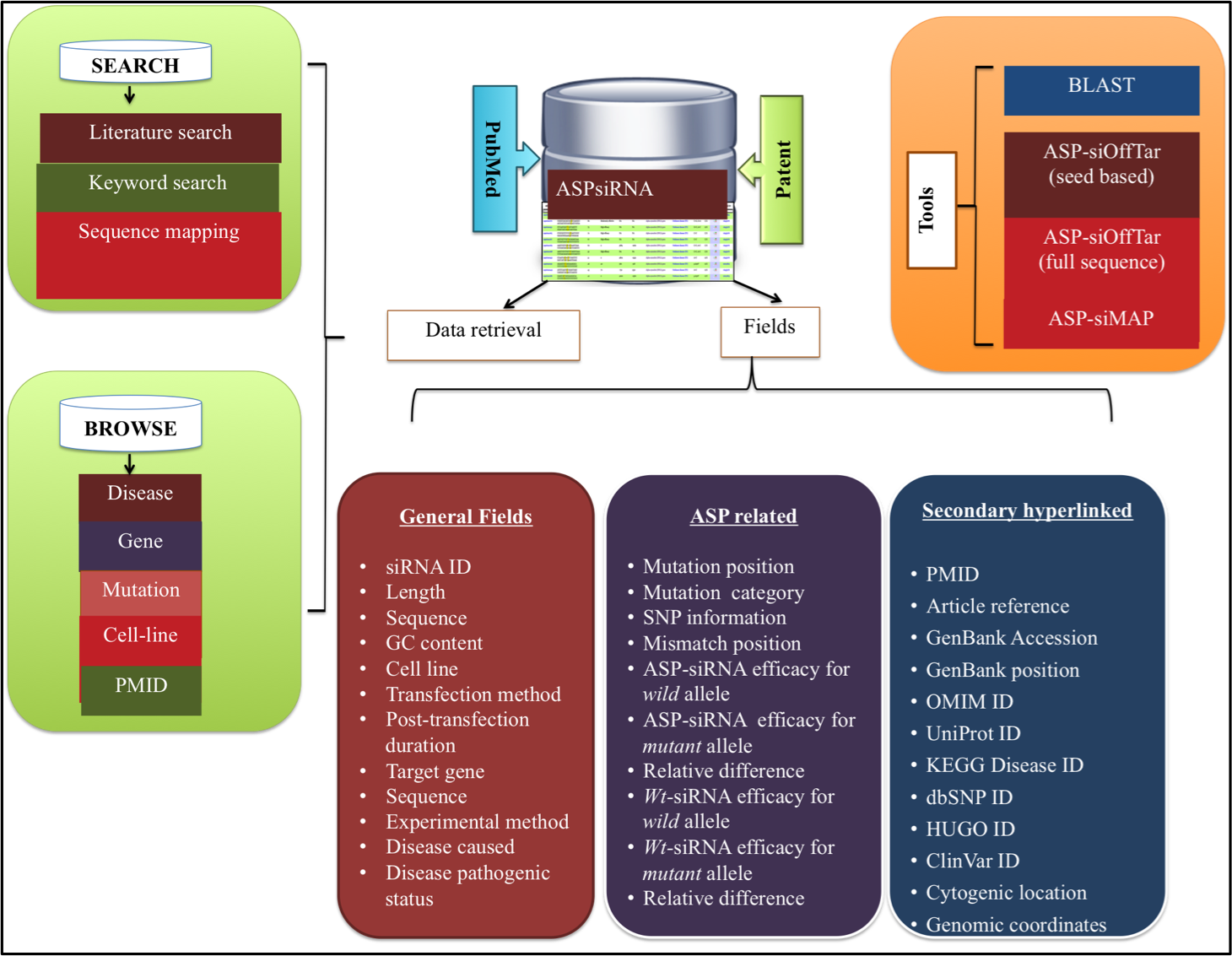
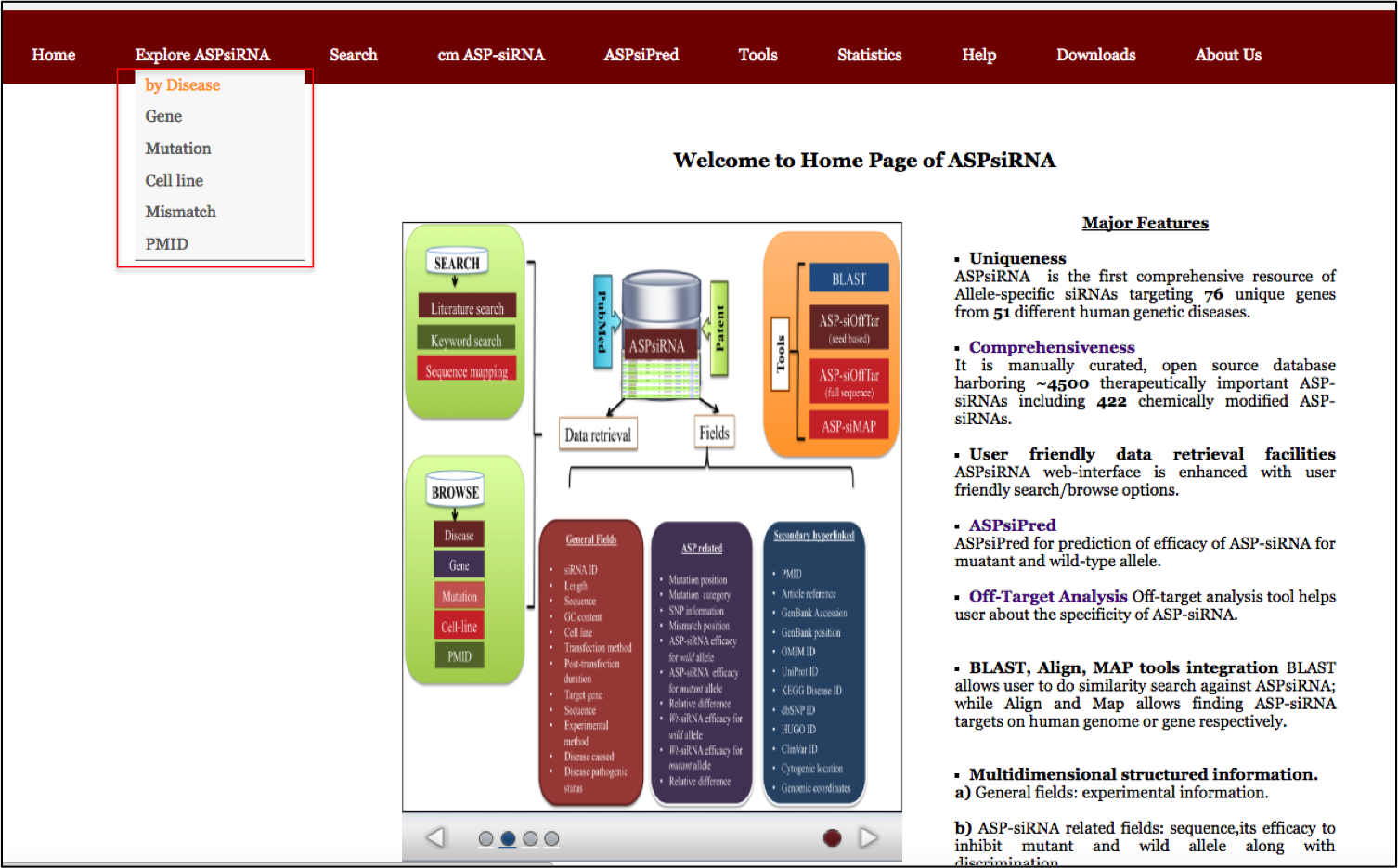
ASPsiRNA (http://crdd.osdd.net/servers/aspsirna/) is the first comprehensive resource in the field of ASP-RNAi, which covers database of therapeutically important ,experimentally validated ASP-siRNAs and prediction of thier inhibitory efficacy. Currently, this compendium covers three components (i) ASPsiDb, an open-source and manually curated database. (ii) ASPsiPred, a two-layered algorithm to predict inhibitory efficacy of ASP-siRNAs for mutant allele (Effmut) by ASPsiPredSVM and wild-type counterpart (Effwild)by ASPsiPredmatrix. (iii) This resource also includes off-target analysis tools like ASP-siOffTar. ASPsiRNA would be immensely helpful for designing and predicting efficacy of ASP-siRNAs for various diseases e.g. in cancer associated SNPs, for treatment of genetic diseases e.g. from currently incurable autosomal dominant to severe sex-linked disorders, in combating viral drug resistance and many more.
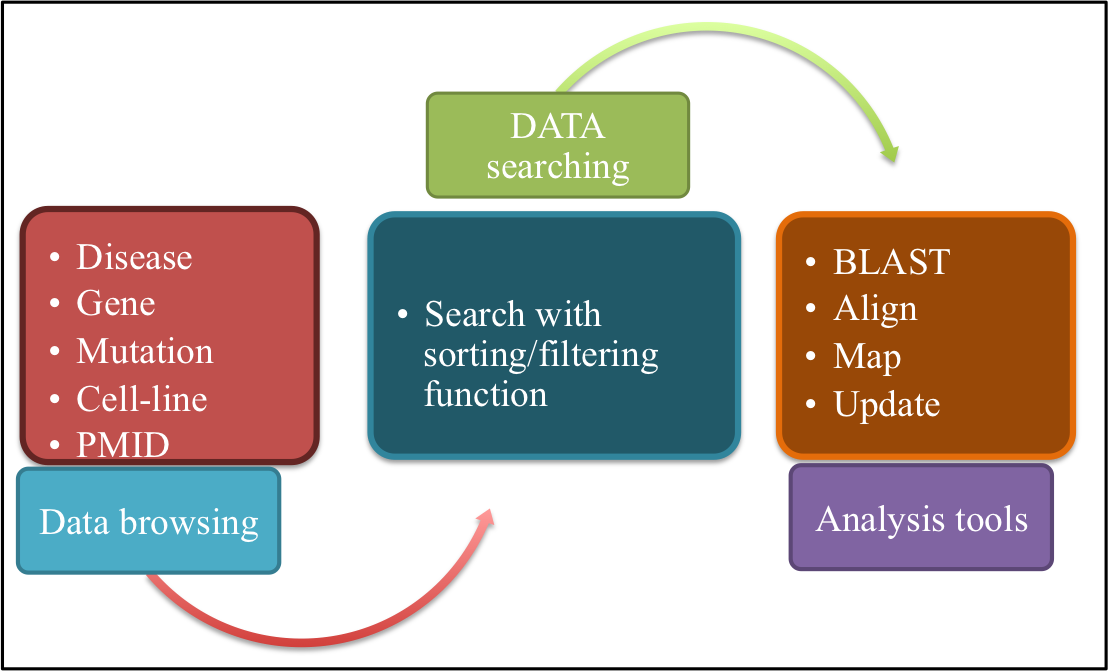
Currently, ASPsiDb contains experimentally verified 4583 ASP-siRNAs targeted against 78 different genes/gene variants from 51 different human disease. These were validtaed on on 45 different cell lines apart from ex-vivo/trangenic mouse models and in-vivo studies. ASPsiDb provides information of ASP-siRNA sequences, mismatch position, efficacy to suppress mutant (Effmut) and wild-type allele (Effwild) along with complete experimental details. It is also enriched with multidimensional genetic and clinical information for numerous variants/mutations. Knowledge about the genetic basis of the disease and clinical information for numerous variants/mutations associated with genes such as mutation position, mutation category and their pathogenic status were collected from OMIM, dbVar, KEGG, and ClinVar. Gene annotations of diverse human genes/diseases were also performed using OMIM, KEGG, UniProt ID, UCSC genome browser; HGNC approved standard name, HUGO ID, NCBI nucleotide ID and GenBank accession and hyperlinked respectively. Database also provides some informative tools for data analysis such as BLAST, ASPsiOffTar and MAP.Database is designed with the aim to increase our understanding of human genetic giseases by mutations and provide maximum useful information to the user; via different tools and external links. It is freely available through this url http://crdd.osdd.net/servers/aspsirna. It comes with enhanced user experience and easy to operate browsing as well as searching with sorting and filtering functionalities. This database has been dedicatedly designed, referring to the current needs of scientific community working exhaustively for RNAi therapeutics development and is anticipated to be useful for them who are seeking a committed repository for ASP-siRNAs.
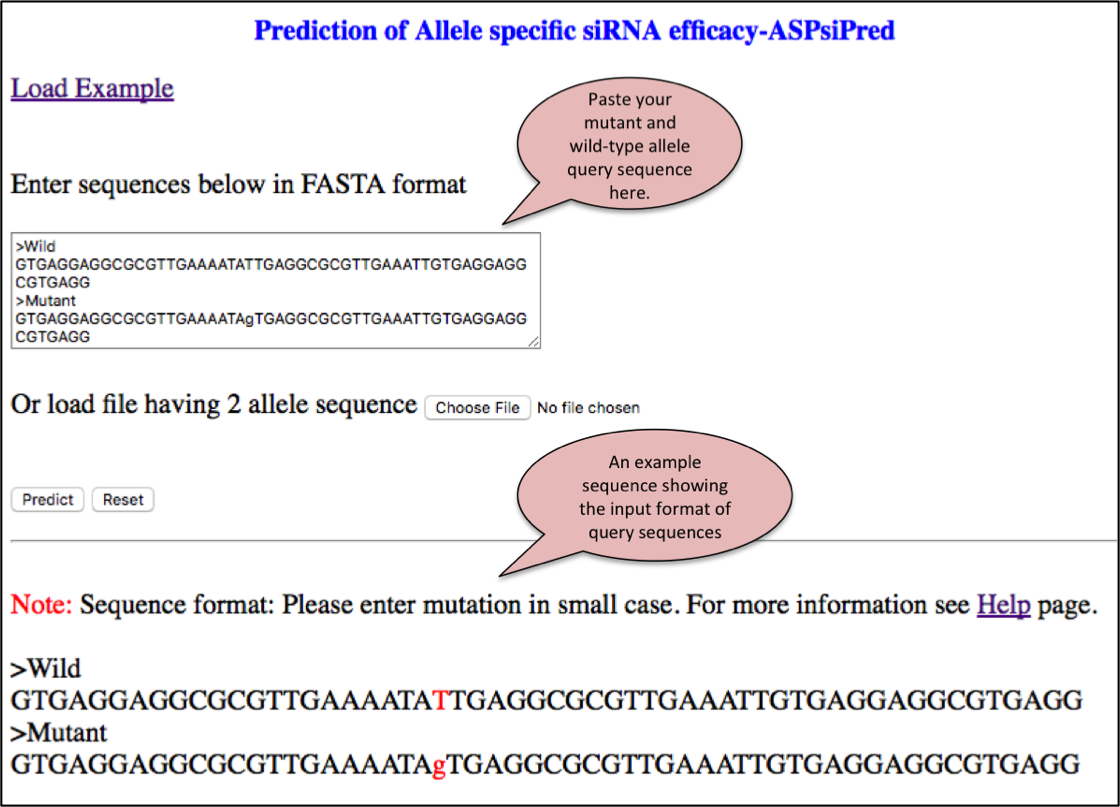
Since, designing of ASP-siRNAs is associated with two efficacies i.e. Effmut and Effwild for mutant and wild-type allele respectively. We have integrated two-layered algorithm in ASPsiPred (ASPsiPredSVM and ASPsiPredMatrix) to predict Effmut and Effwild respectively.
In ASPsiPredSVM, we have employed a dataset of 922 ASP-siRNAs with experimentally validated quantitative efficacies. These siRNAs were divided into training (T737) and validation (V-185 datasets. Various siRNA features like mono- to penta-nucleotide frequencies, binary pattern and their hybrid combinations were used for model development. Further, to predict Effwild, a 2D-matrix termed as ASPsiPredmatrix was constructed from experimentally validated rule-based studies describing effect of siRNA: mRNA residue clash at 19-different locations.
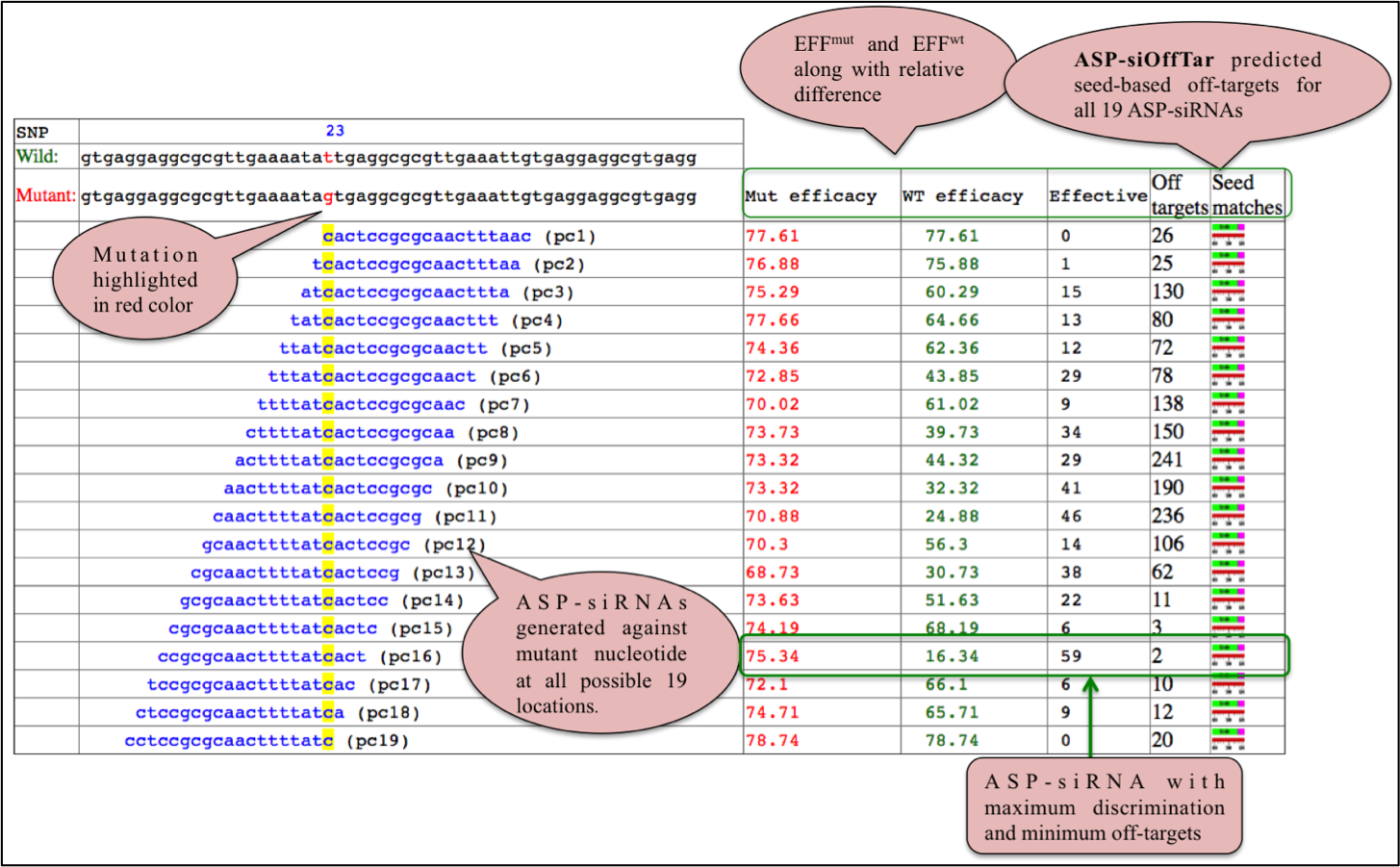
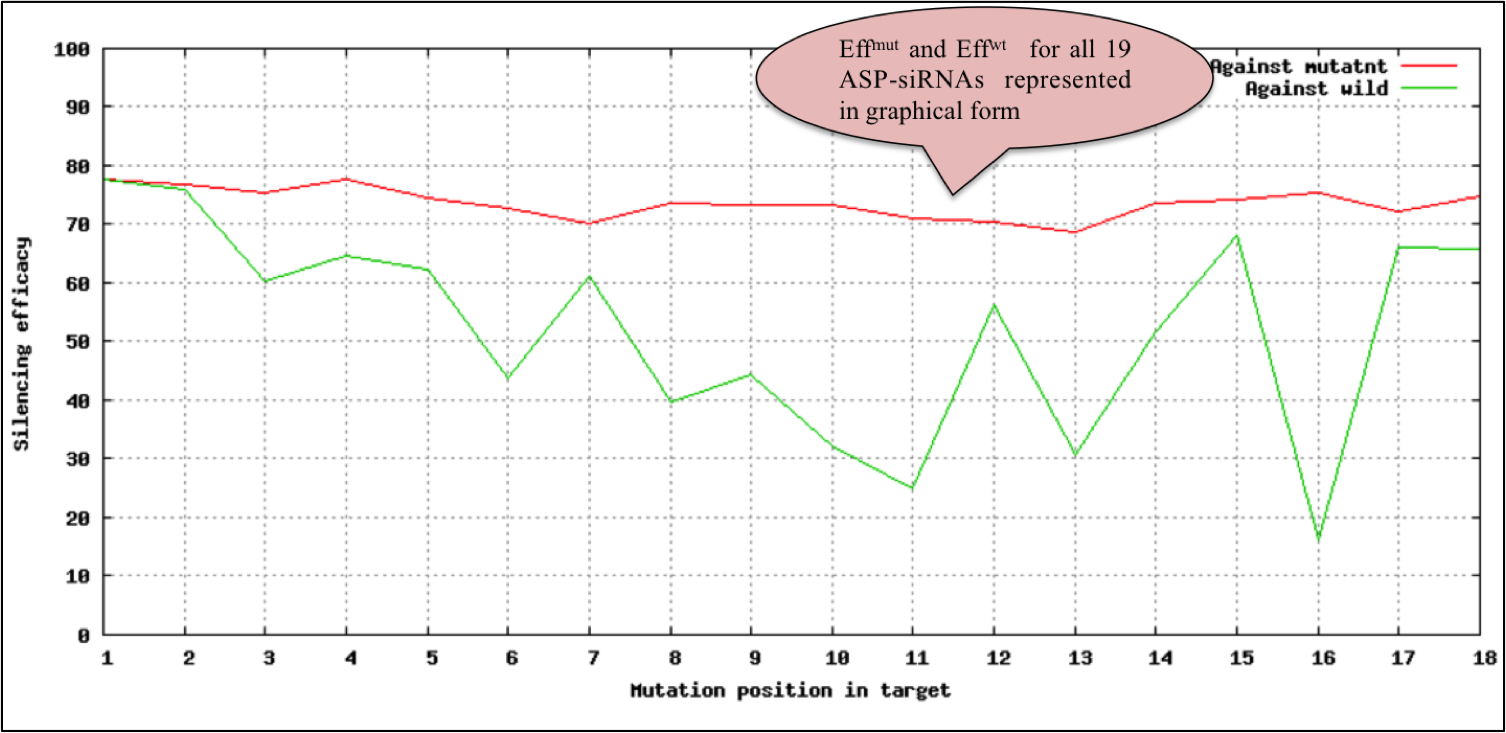
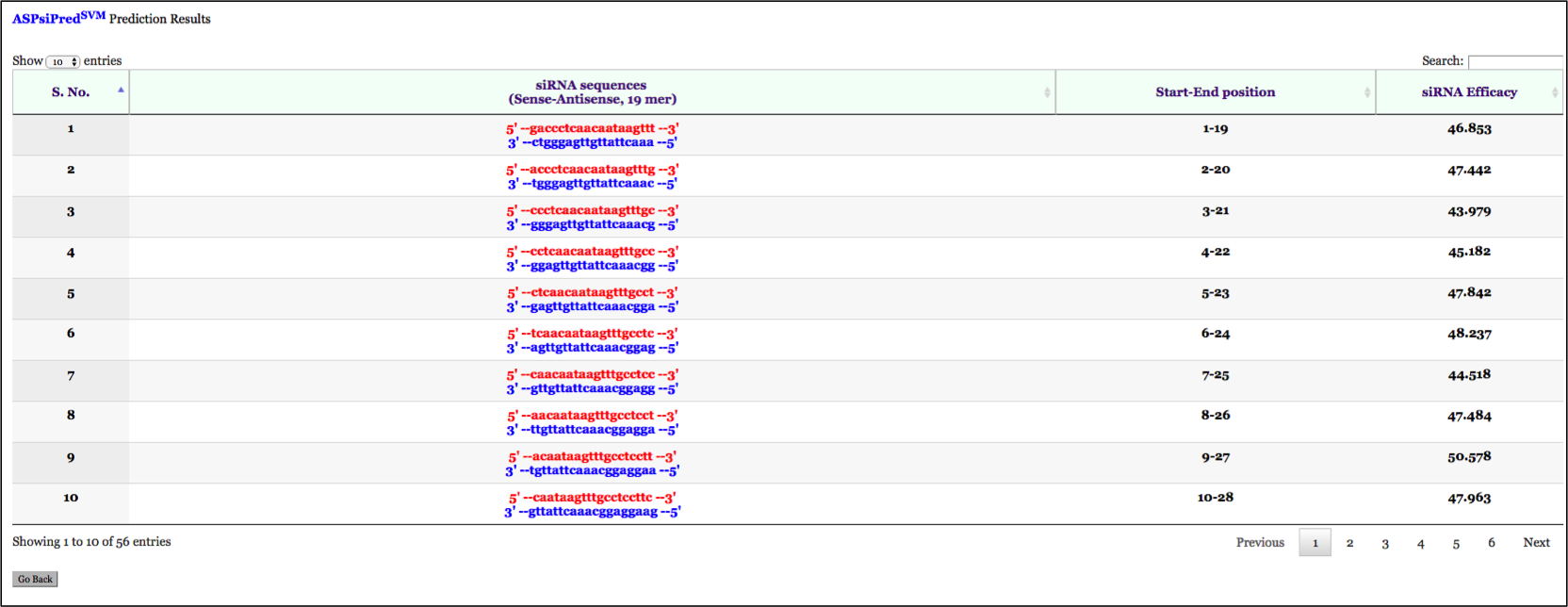
User can submit mutant and wild-type allele along with mutated base in lower case.This tool allows the efficacy prediction of 19-mer ASP-siRNAs fully complementary with mutant allele while having one mismatch with wild-type allele at all possible 19 locations. Efficacy to suppress mutant (Effmut)as well as wild-type allele (Effwild) is listed in the form of table. A graphical representation to assess the relative discrimination of each ASP-siRNA is also provided below the table.

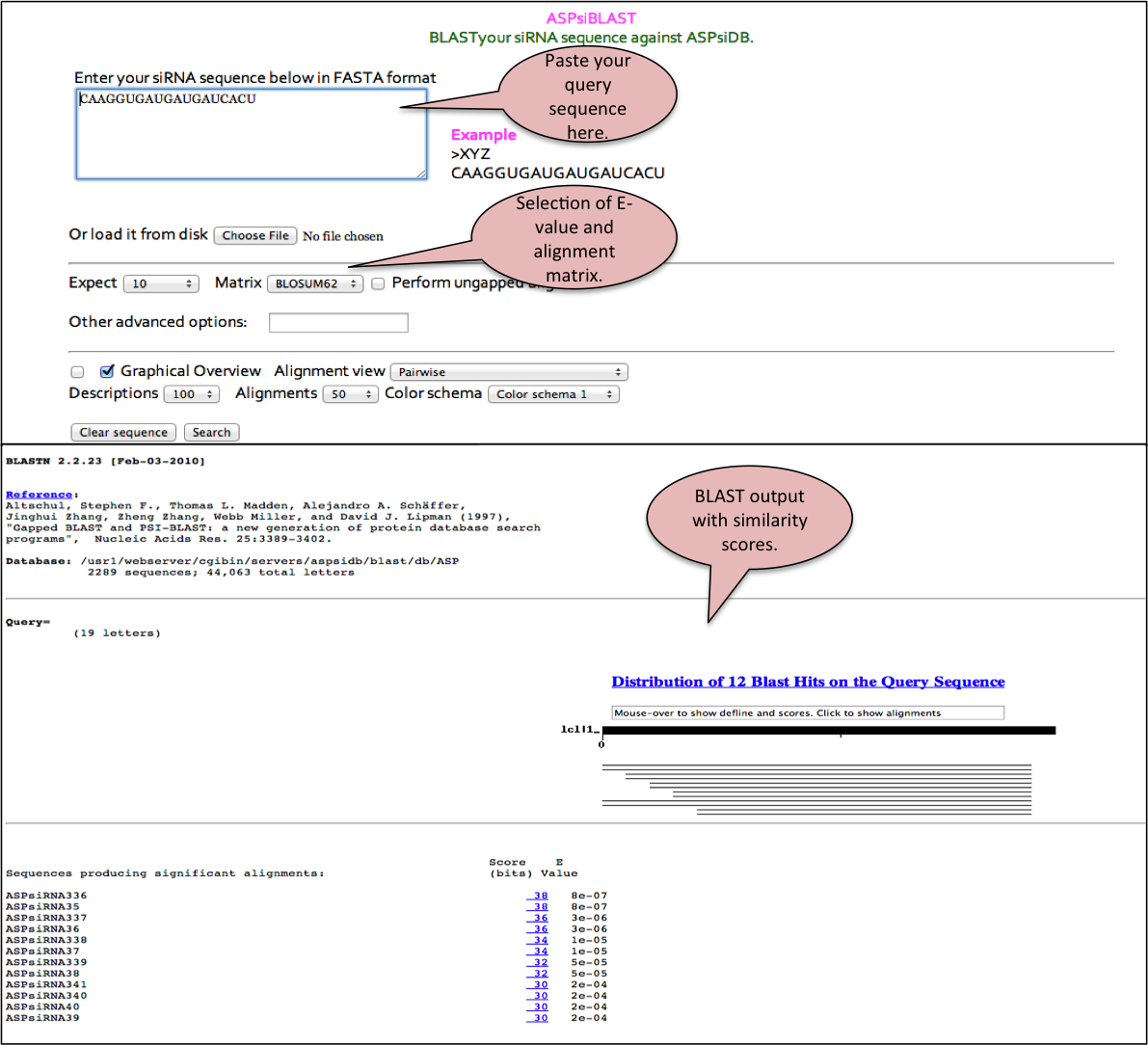
ASPsiBlast
This tool allows alignment of a user provided siRNA sequence against all the ASP-siRNA sequences available in our database. This helps the user to confirm whether a given siRNA sequence or similar one has already been reported or not.
The output is given in the standard format with the blast score and e-value. The alignment is shown for ASP-siRNAS found to be identical or similar in the database. The output can be formatted based on the options provided by user.
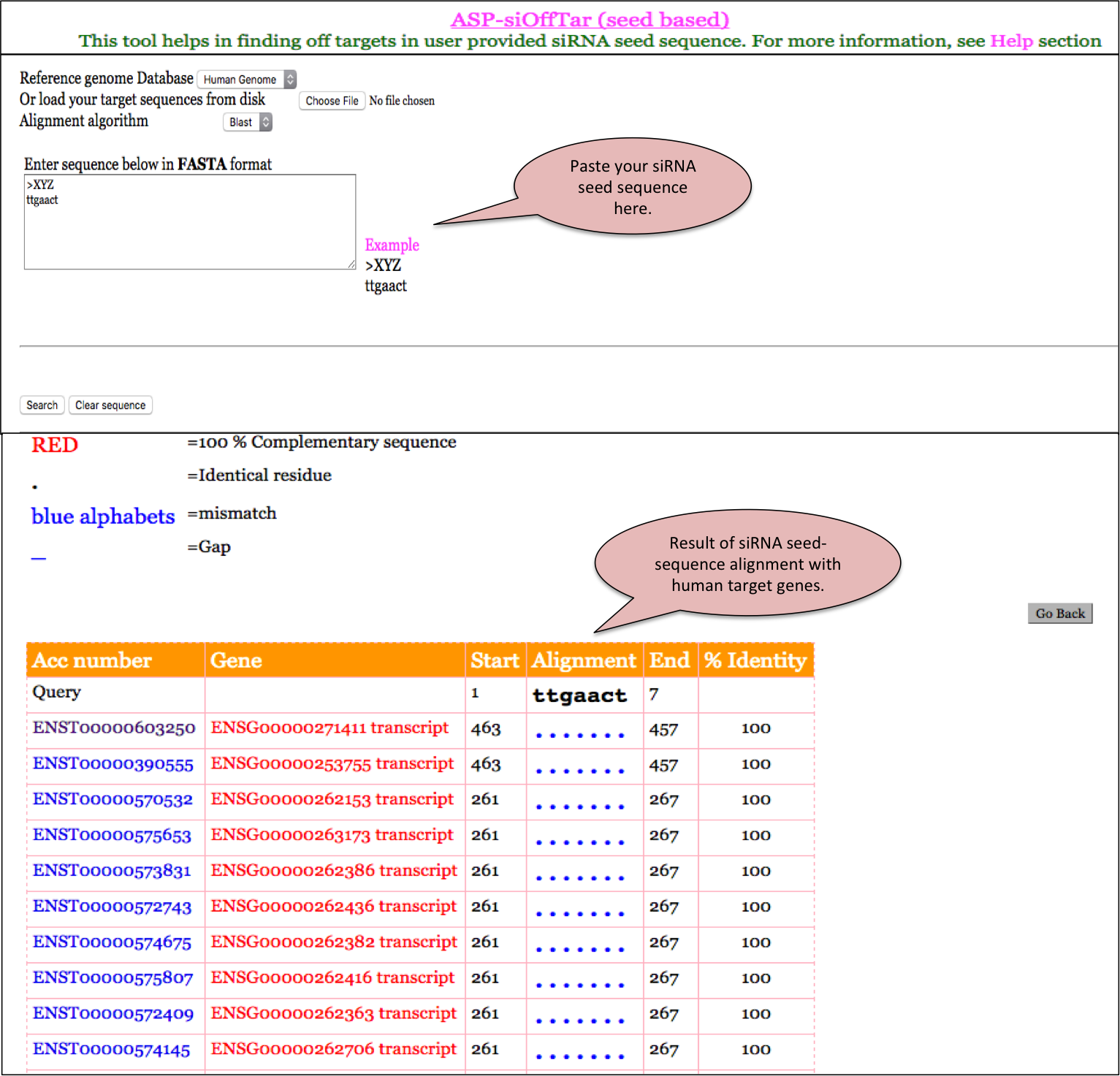
ASP-siOffTar (seed-based)
ASPsiAlign allows alignment of a user provided aligns the user given siRNA sequence on the human reference genome.
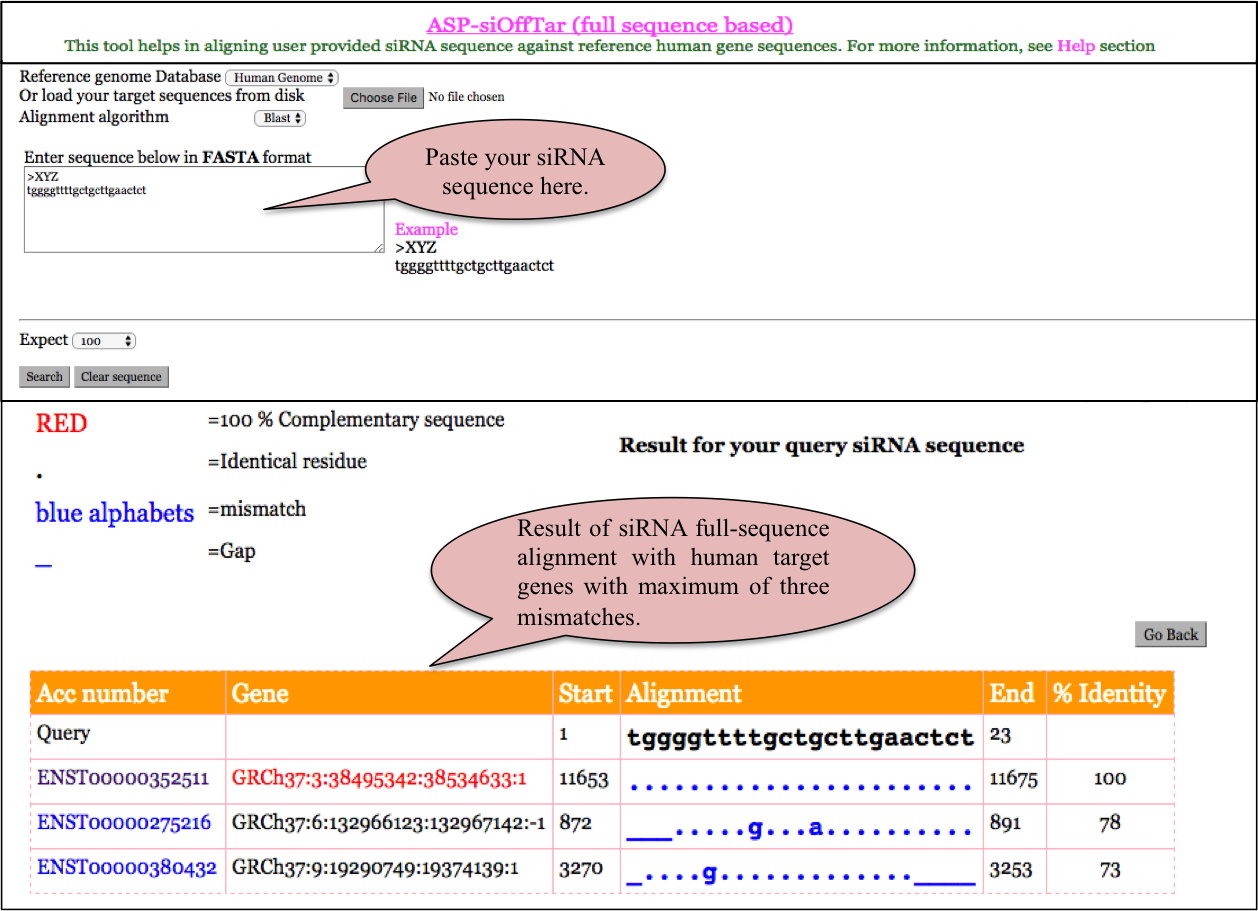
ASP-siOffTar (full-sequence based)
ASPsiAlign allows alignment of a user provided aligns the user given siRNA sequence on the human reference genome.
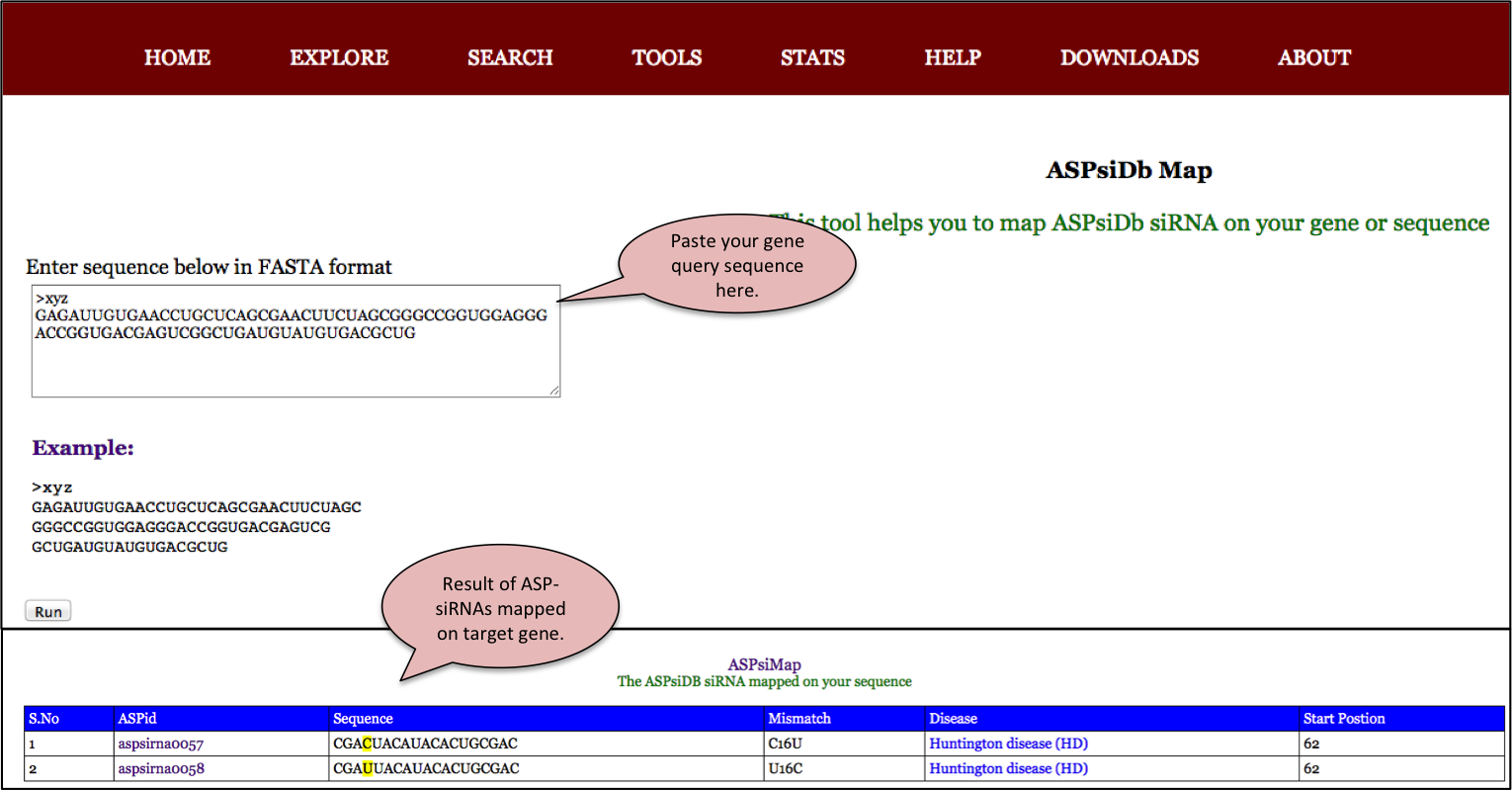
ASPsiRNA Map
This tool can be use to map the ASP-siRNAs in the database against the user given target gene sequence. Therefore it can be helpful to use these siRNAs for the suppression of desired target gene sequence.
User can use search facility in the following two ways:
1) User can perform a simple search by giving a keyword to the dialog box provided on the seacrh page and click submit.
Input: Here you can enter your desired "Keyword" in the dialog box and can search against all fields by specifying 'All' radio button or any of the 16 representative fields of the entire database categorized in three parts as
ASPsiRNA details like ASP-siRNA ID,ASP-siRNA name, its antisense sequence, mismatch in siRNA, ASP-siRNA percentage efficacy against mutant allele, relative difference.Target gene/disease details like target gene, target mutation, mutation category/variant, SNP, disease and pathogenic status of mutation pertaining to that disease. Important Experimental details like cell line, experimaental technique used to determine efficacy/expression of ASP-siRNAs like dual luciferase reporter assay, PMID, Author.
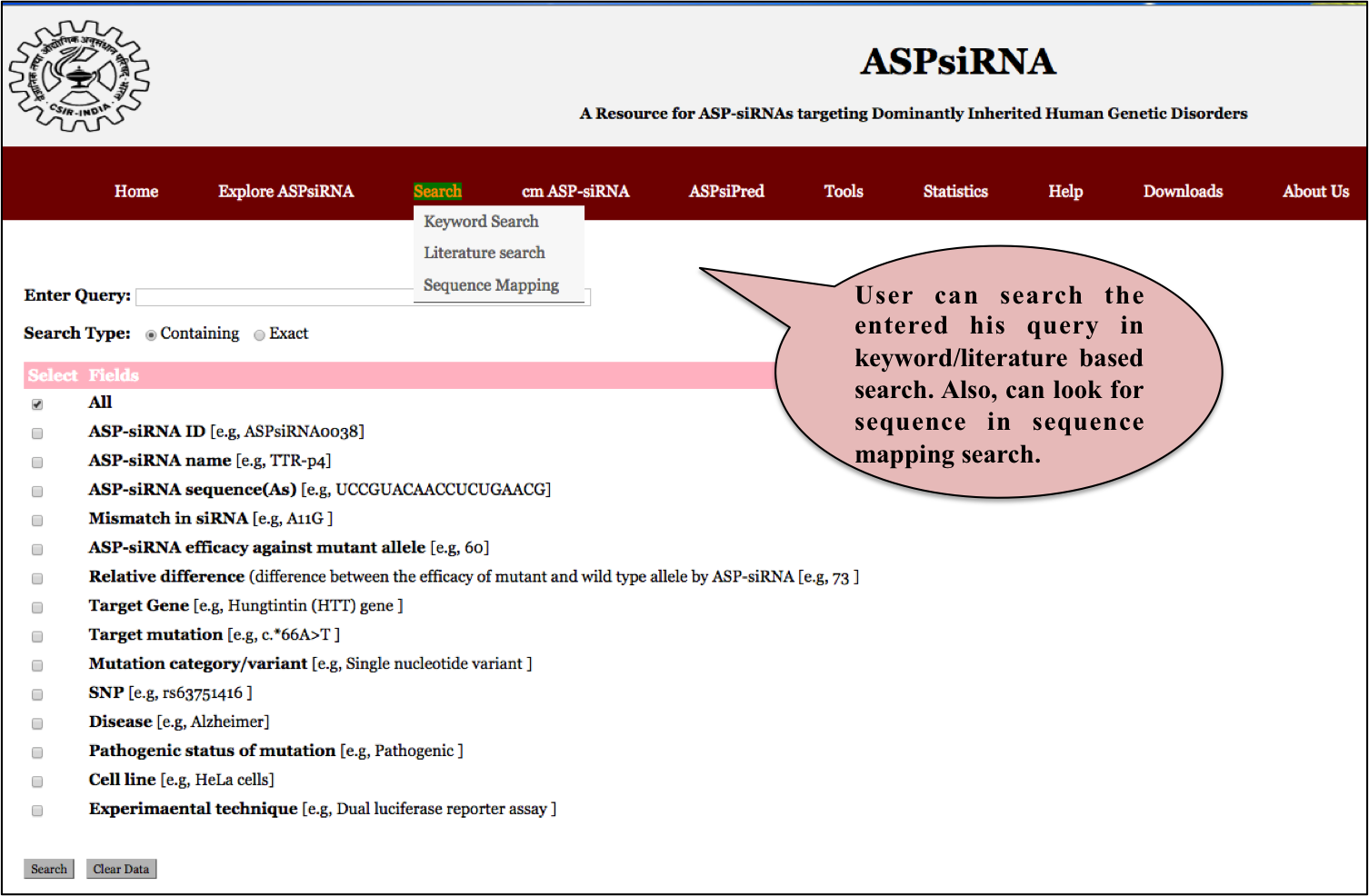
Default option is All radio button, which searches against all the fields in the database. Examples have been provided corresponding to each field which can be used as sample queries. Besides the option to choose the field, search type allows to retrieve either an exact match or the match containing the query. Clicking on clear data button clears the data after your search.

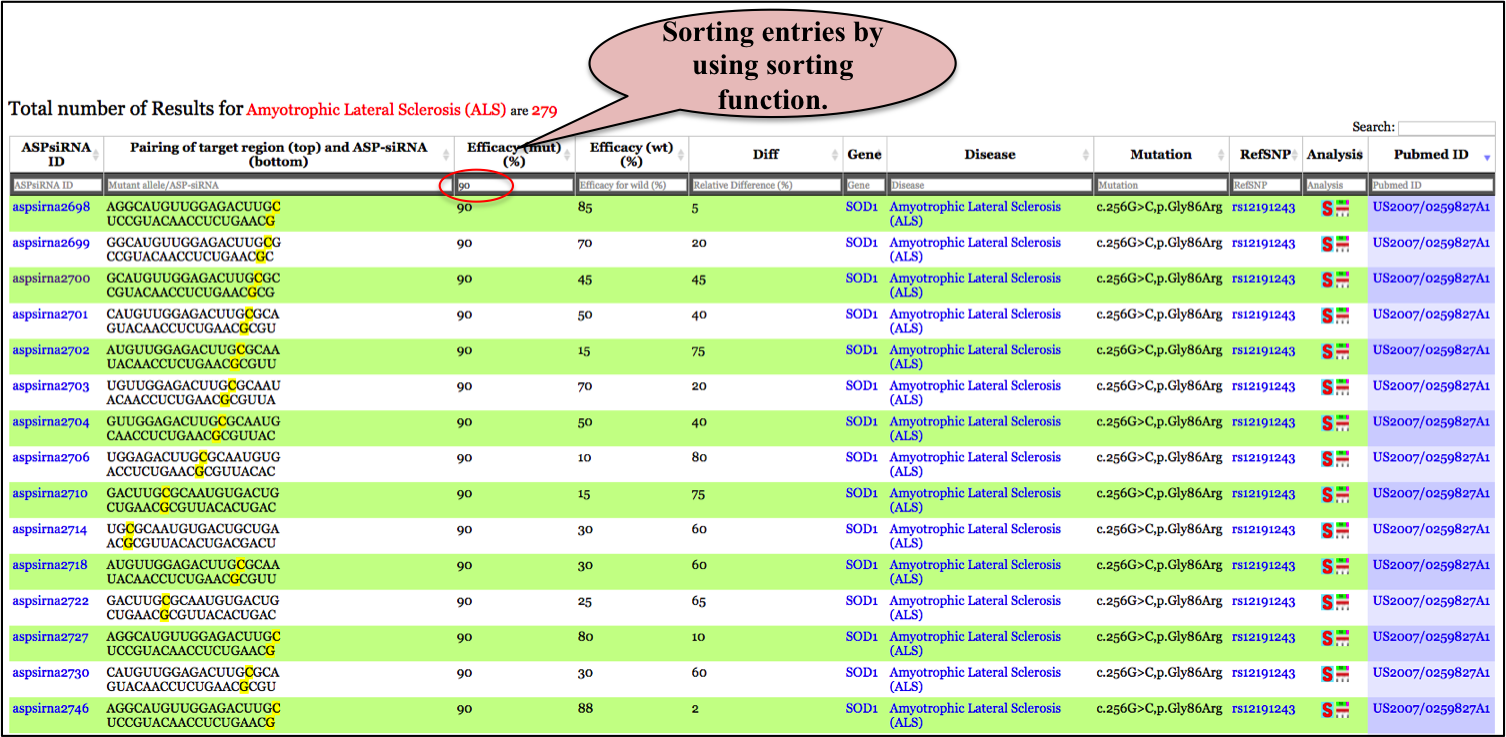
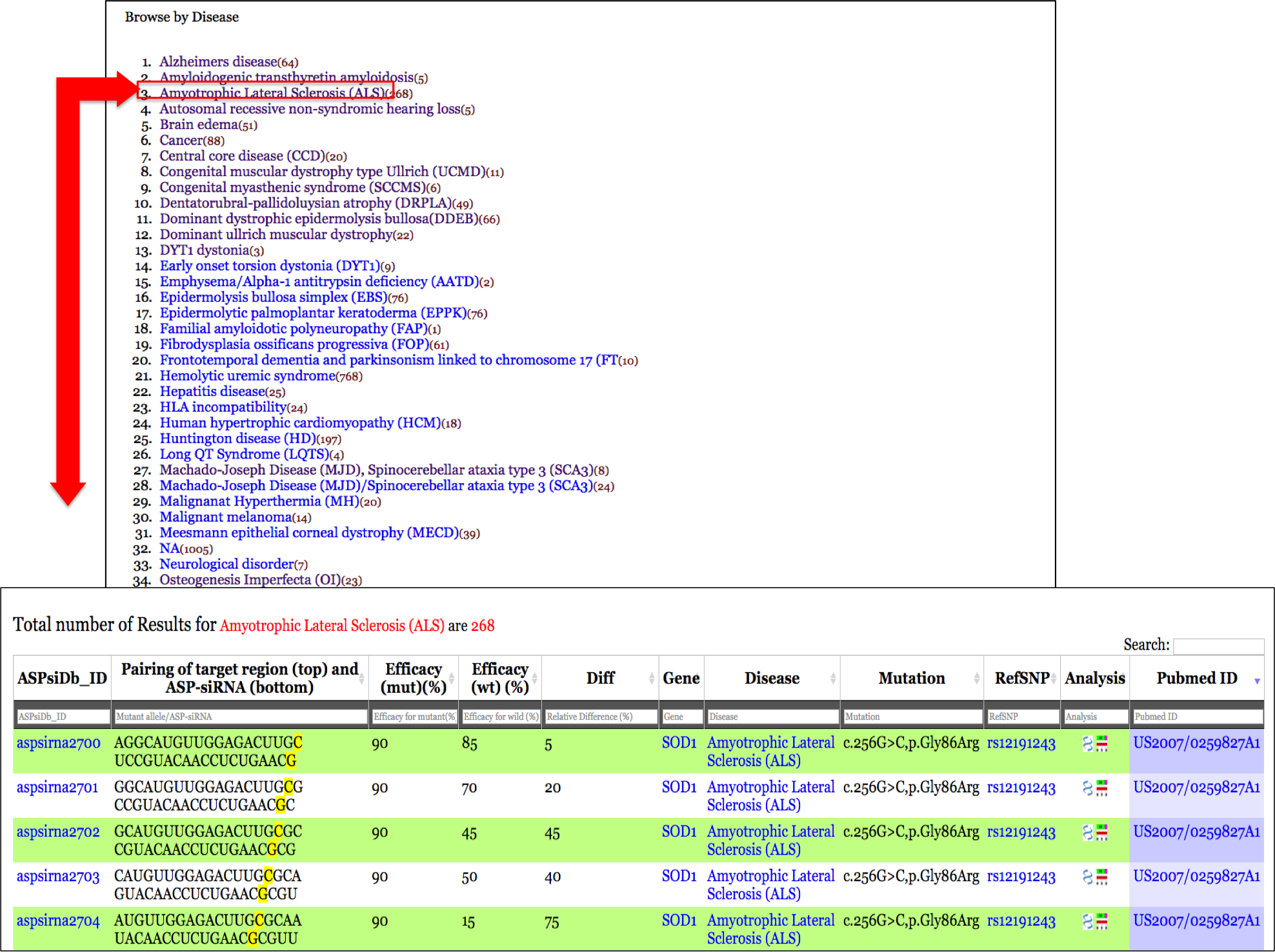
Output: The results obtained from the keyword "Amyotrophic Lateral Sclerosis" search displays a total of 268 entries at top of main table. Tha main table harboring data related to ALS disease displays 11 representative fields as
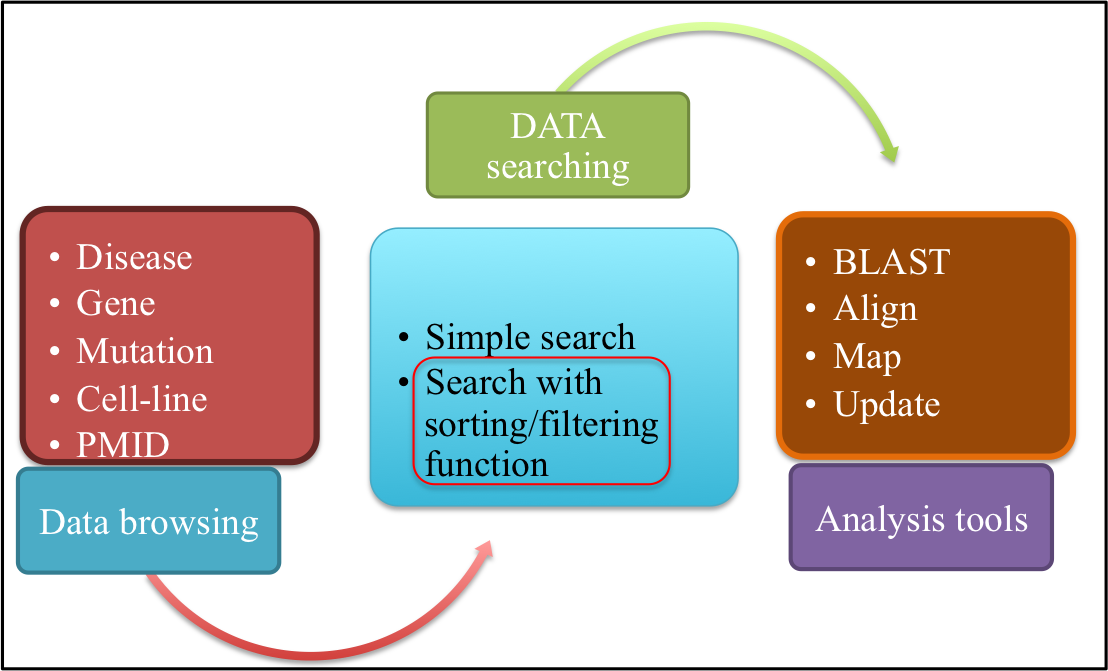
You can sort/filter the data generated by simple search by typing in the radio buttons provided at the top of every field in the main table.For eg: by performing simple search for disease "Amyotrophic Lateral Sclerosis" displays 268 entries; but suppose if a user wants all ASP-siRNAs targted against ALS i.e Amyotrophic Lateral Sclerosis disease AND having percentage efficacy of 90%. User can enter 90 in the radio button provided above the Mutant efficacy(%) field. The result displays 20 ASP-siRNA entries causing Amyotrophic lateral sclerosis AND having percentage Mutant efficacy 90%. Output is depeicted in figure below showing 20 entries filtered from 268 entries.
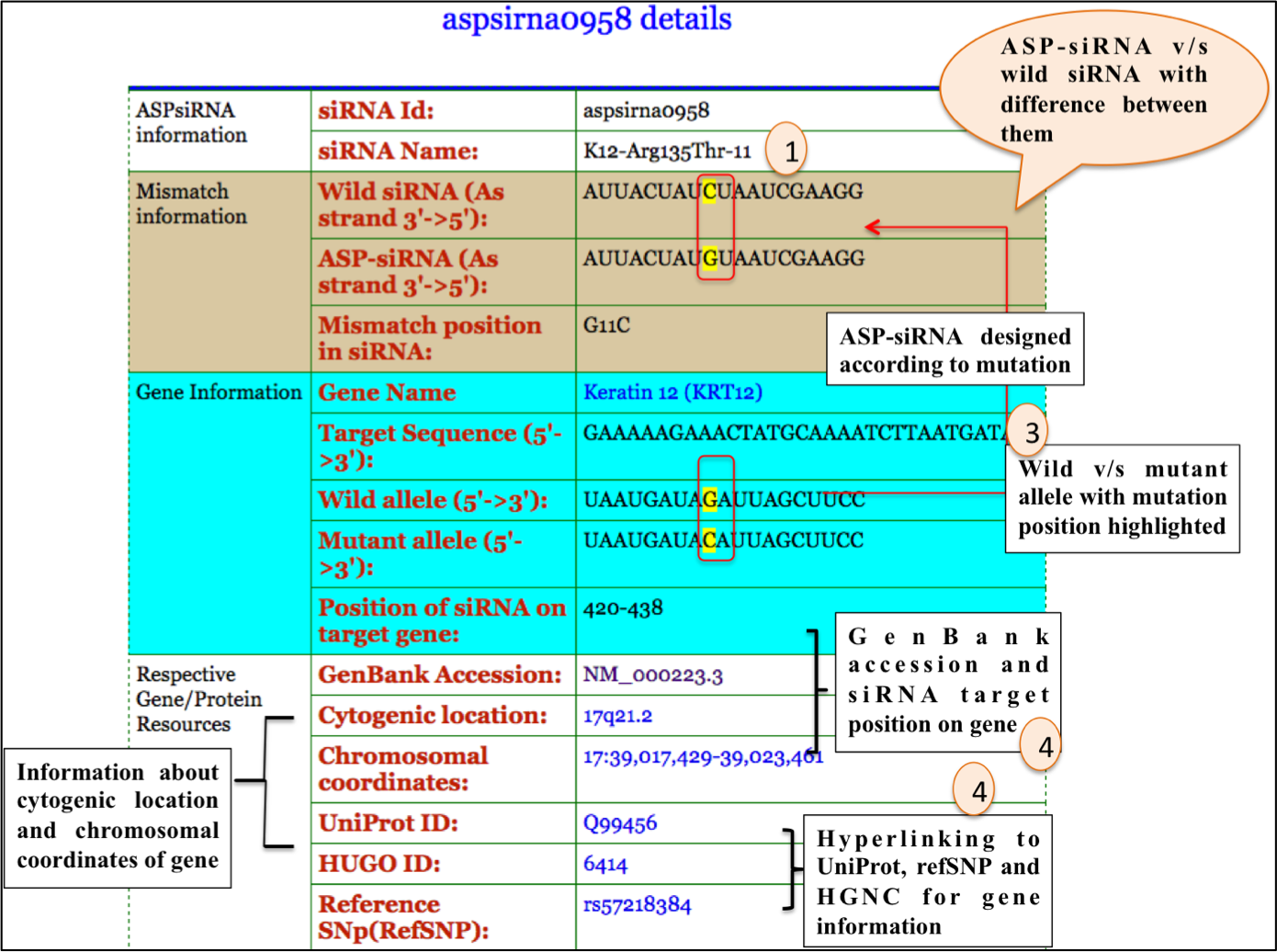
Upon clicking individual ASP-siRNA ID; user can get comprehensive information about particular
1. ASP-siRNA
2. Detailed information about Gene target by ASP-siRNA.
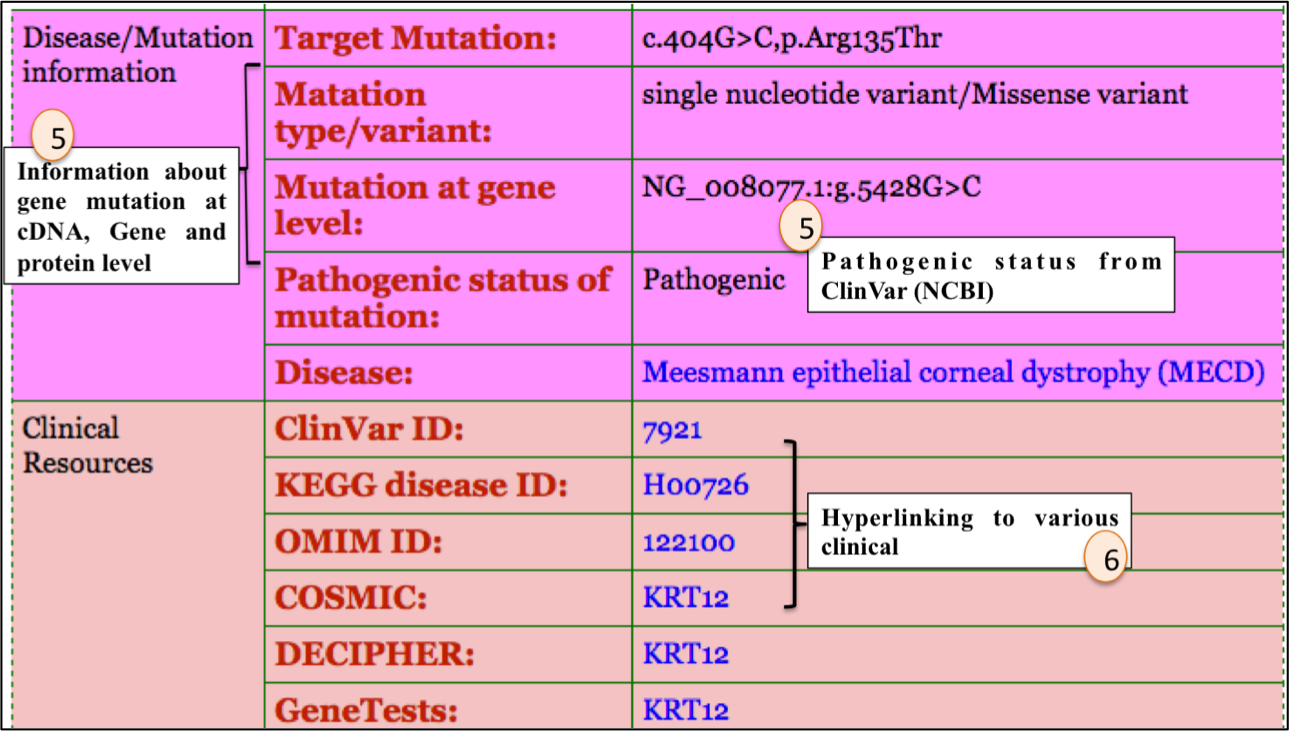
3. Information about disease and mutation causing respective disease.
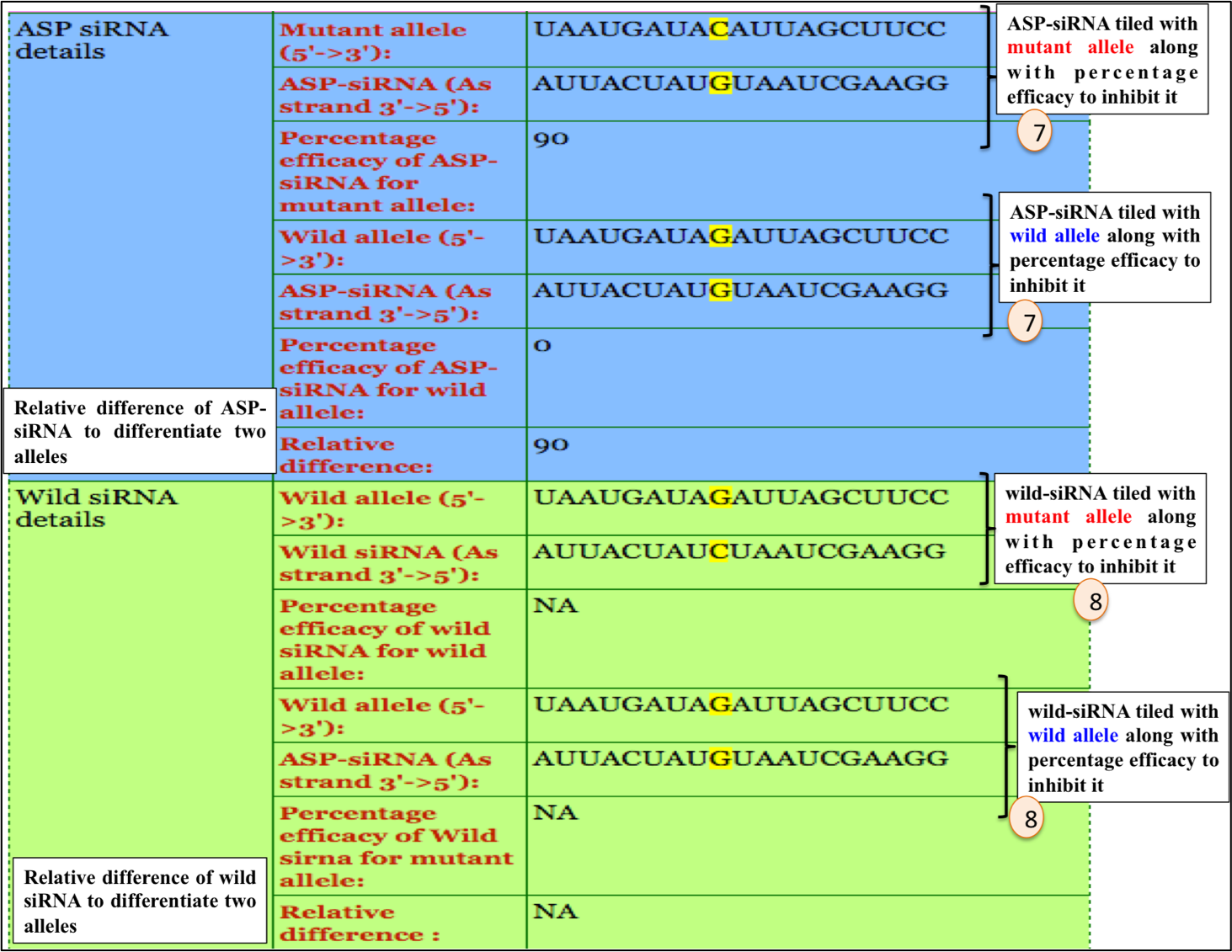
4. ASP-siRNA efficacy details.
5. Wild siRNA efficacy details.
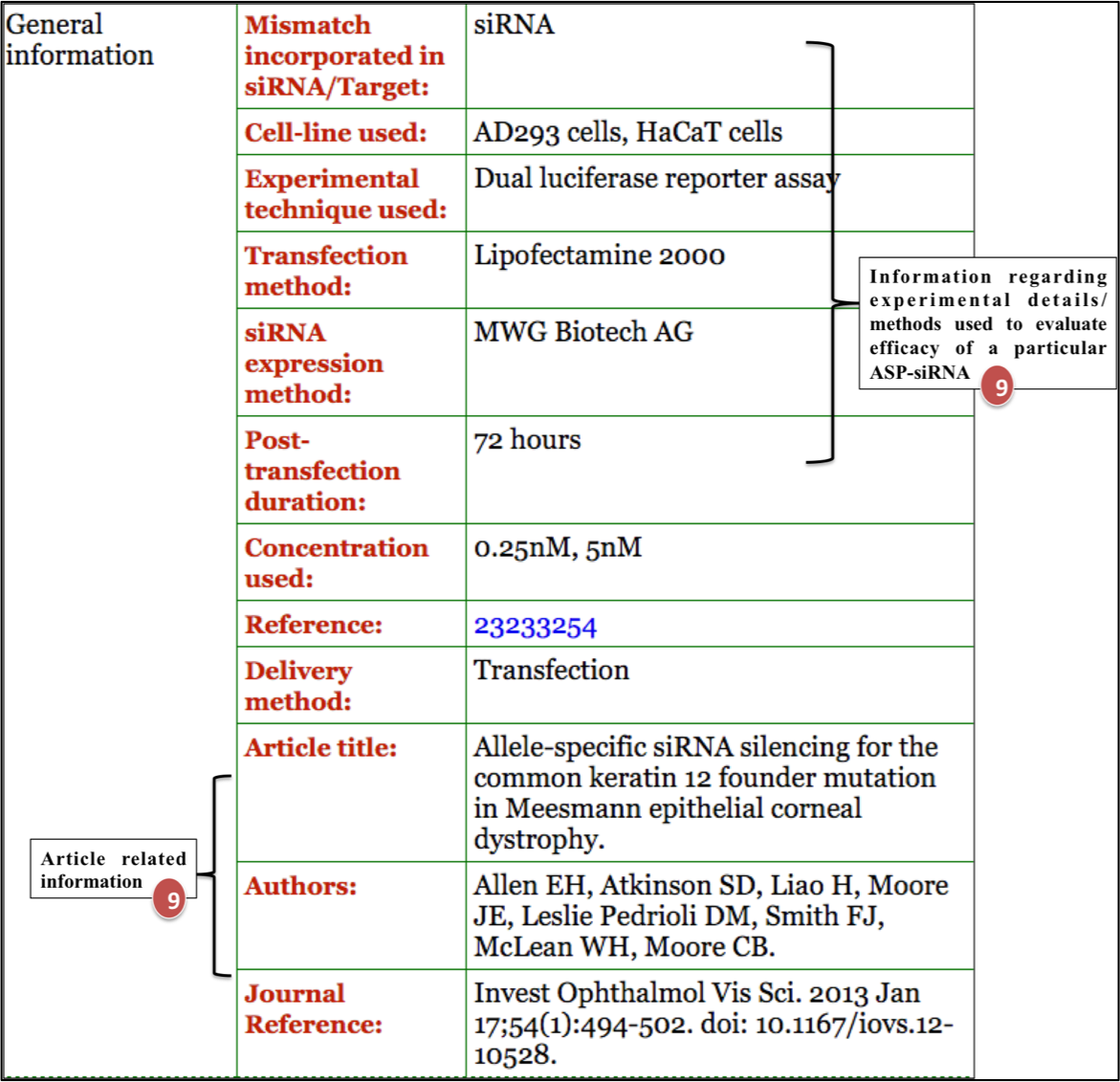
6. General information.
For example: ASPsiRNA0960; which is designed to target Meesmann epithelial corneal dystrophy (MECD). Upon clicking ASPsiRNA0960; detailed information is displayed as depicted in figures below:
ASPsiRNA contains highly curated and updated experimental data on ASP-siRNAs. It will be updated on annual basis depending upon the number of new studies in the field.