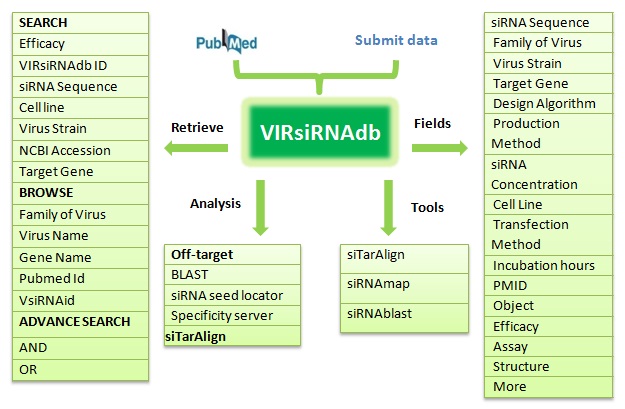
Viral siRNA Database (VIRsiRNAdb) is a structured collection of experimentally validated siRNAs reported in the scientific literature targeting diverse genes of important human viruses. The curation efforts to date has concentrated mainly on viruses known to be associated with infectious diseases in humans. Currently the database provides experimental information of 1358 siRNAs, mined from relevant Pubmed artcles, which includes siRNA sequence, virus subtype, target gene, GenBank accession, design algorithim, cell type, and efficacy. Further, wherever available, extended information regarding alternative efficacy assays has also been provided. VIRsiRNAdb also allows the users to take advantage of useful tools like siRNAblast, siRNAmap and siTarAlign. As a result our database will become increasingly useful for developing siRNA design tools. Additionally this database will help the user to confirm whether a given siRNA sequence or similar one has already been reported or not and also what experimental conditions were used and which genomic segments are more indispensable for different human viruses.
Some other siRNA databases which may be useful to you are listed below:
|
Database |
Year |
siRNA |
Working URL |
|
HIVsirDB |
2011 |
HIV |
|
|
siRNAdb |
2005 |
Human |
|
|
HuSiDa |
2005 |
Human |
|
|
siRecords |
2006 |
Mammalian |
|
|
siRecords |
2009 |
Mammalian |
|
|
DSTHO |
2006 |
Oncogenes |
Search
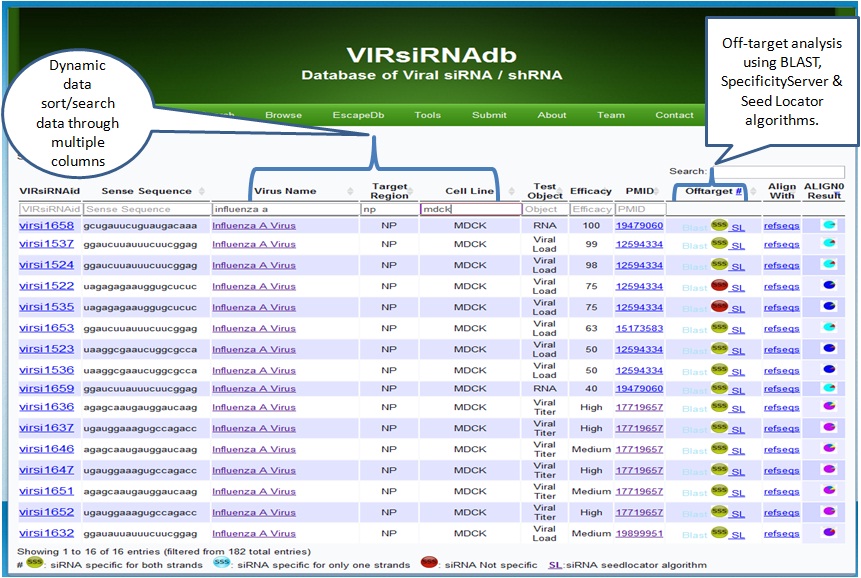
The user may perform a quick search based on VIRsiRNAdb ID, Target Site, Sequence, Virus name, Virus Strain,NCBI Accession,Target Gene, and siRNA efficay level from the database search page.
Advance search
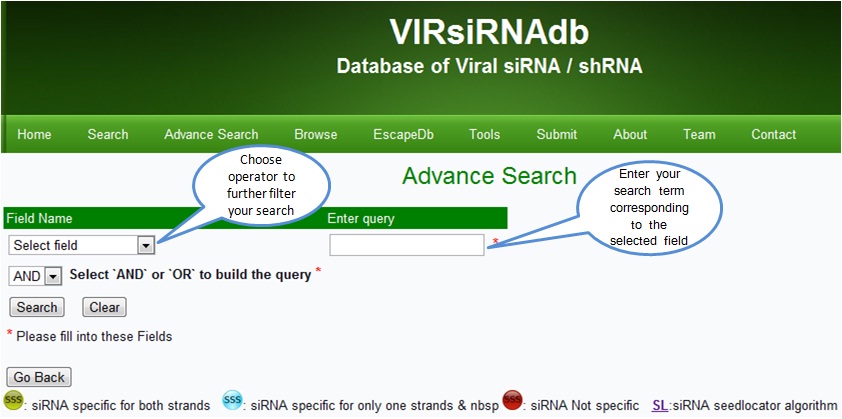
Advanced Search page allows for more flexible queries based on criteria such as the viral data set of interest using logical operators (AND, OR). The search returns a list of siRNAs, and by clicking on the gene/virus name the database will present a summary of the record for the selected gene/virus. These options enable the user to readily identify suitable siRNA related data.
Browse
External links pointing to the GenBank accession of the siRNA sequence, Pubmed ID of the Reference paper and International Committee on Taxonomy of Viruses (ICTV) have been placed appropriately for each record in the database.

Browse by Virus name
Tools
Viral siRNA database allows the users to take advantage of useful tools like siTarAlign, VIRsiRNAblast and siRNAmap.
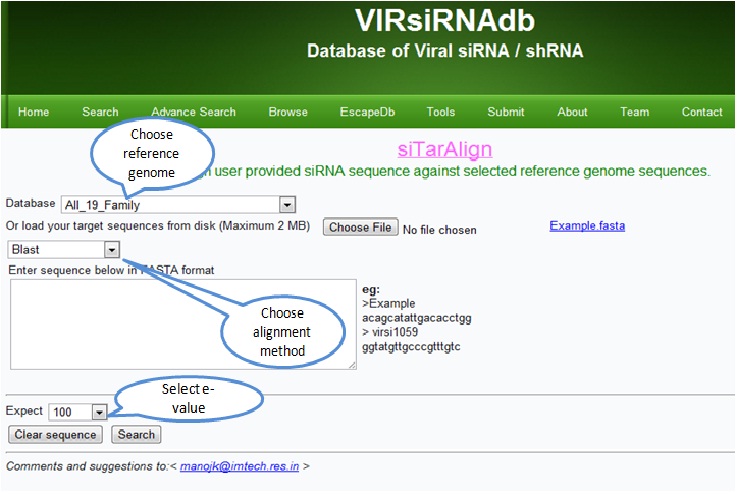
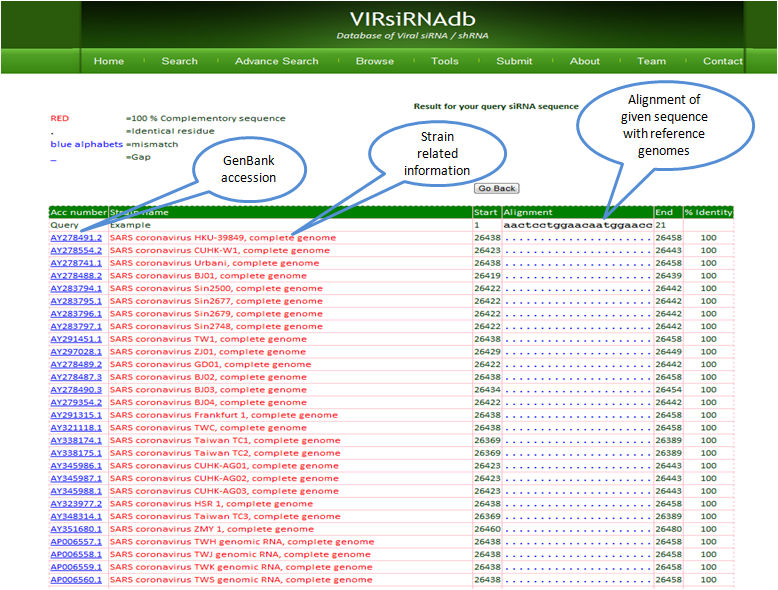
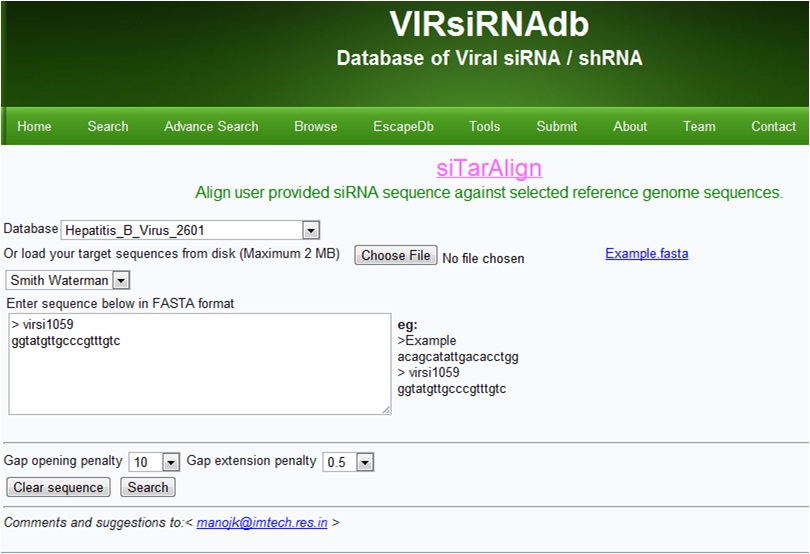
siTarAlign
siTarAlign aligns the given siRNA sequence in target reference sequences databases of various viruses .Users can also upload their reference sequences (Maximum upto 2MB).
VIRsiRNAblast
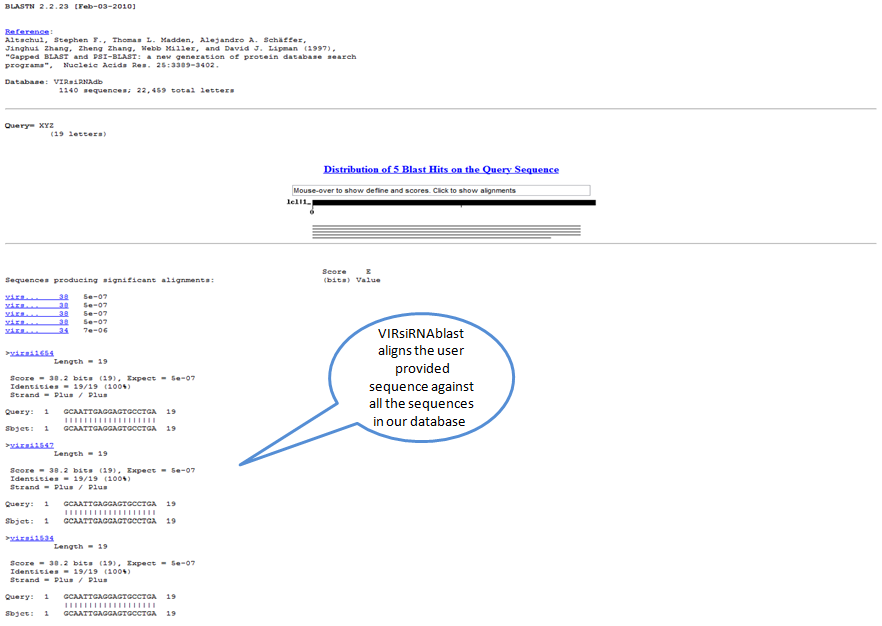
The 'BLAST tool' allows alignment of a given sequence against all the sequences in our database. This may help the user to confirm whether a given siRNA sequence or similar one has already been reported or not.
Output of VIRsiRNAblast:
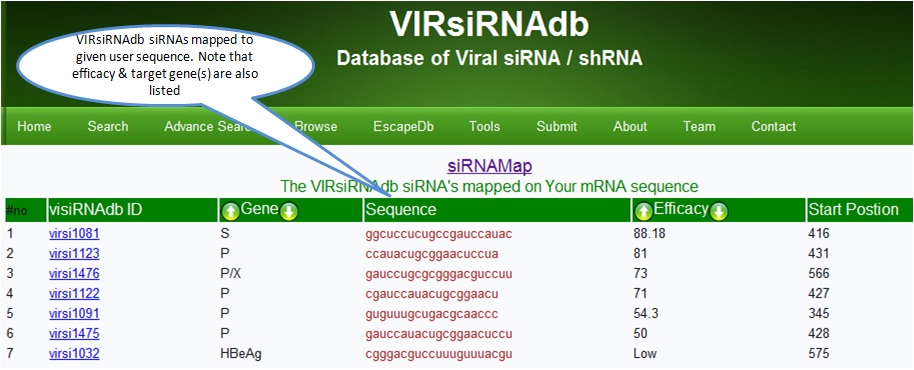
siRNAmap
The 'siRNAmap' maps the VIRsiRNAdb siRNA's on the user nucleotide sequnece .Provide the list of siRNA from database which are complementory to the user provided sequence.
Output of siRNAmap:
Submit
Authors of publications reporting RNA interference involving viral genes are encouraged to submit the data directly to Viral siRNA database. For this purpose, a web form for data submission is provided.
Update
Regular update of the database and inclusion of new viruses is one of our main priority.
FAQ
What is VIRsiRNAdb?
Viral siRNA Database is a structured collection of experimentally validated siRNAs reported in the scientific literature targeting diverse genes of important human viruses.
What are the fields in VIRsiRNAdb?
(1) siRNA sequence, (2) family of virus, (3) virus subtype, (4) target gene, (5) siRNA location, (6) GenBank accession, (7) design algorithim, (8) production method, (9) siRNA concentration, (10) cell type, (11) transfection method, (12) incubation hours, (13) PMID, (14) Object used (i.e. mRNA, protein, virus load etc), (15) efficacy, (16) efficacy assay (e.g. Western blot, PCR, plaque number, ELISA) & (17) references.
What are the tools in VIRsiRNAdb?
Viral siRNA database allows the users to take advantage of useful tools like siRNA BLAST, siRNA Map and siRNA Align.
What does the BLAST tool do?
The 'BLAST tool' allows alignment of a given sequence against all the sequences in our database. This may help the user to confirm whether a given siRNA sequence or similar one has already been reported or not.
What is the use of siTarAlign?
siTarAlign aligns the siRNA target in user defined reference viral genome.
What is the use of siRNAmap?
The siRNAmap lists possible siRNA's that can target the user sequence region(s). This enables alignment of Viral siRNA database siRNA sequences on the user sequence. The output shows a list of siRNA's that can target the user sequence along with the target position and efficacy of the siRNA.
How to use search in VIRsiRNAdb?
The user may perform a quick search based on VIRsiRNAdb ID, Target Site, Sequence, Virus name, Virus Strain,NCBI Accession,Target Gene, and siRNA efficay level from the database search page.
How to use advanced search in VIRsiRNAdb?
‘Advanced Search page’ allows for more flexible queries based on criteria such as the viral data set of interest using logical operators (AND, OR). The search returns a list of siRNAs, and by clicking on the gene/virus name the database will present a summary of the record for the selected gene/virus. These options enable the user to readily identify suitable siRNA related data.
How many entries are there in VIRsiRNAdb?
Currently the database provides experimental information of 1358 siRNAs.
How many extended entries are there in VIRsiRNAdb?
Currently the database provides extended information of 300 siRNAs.
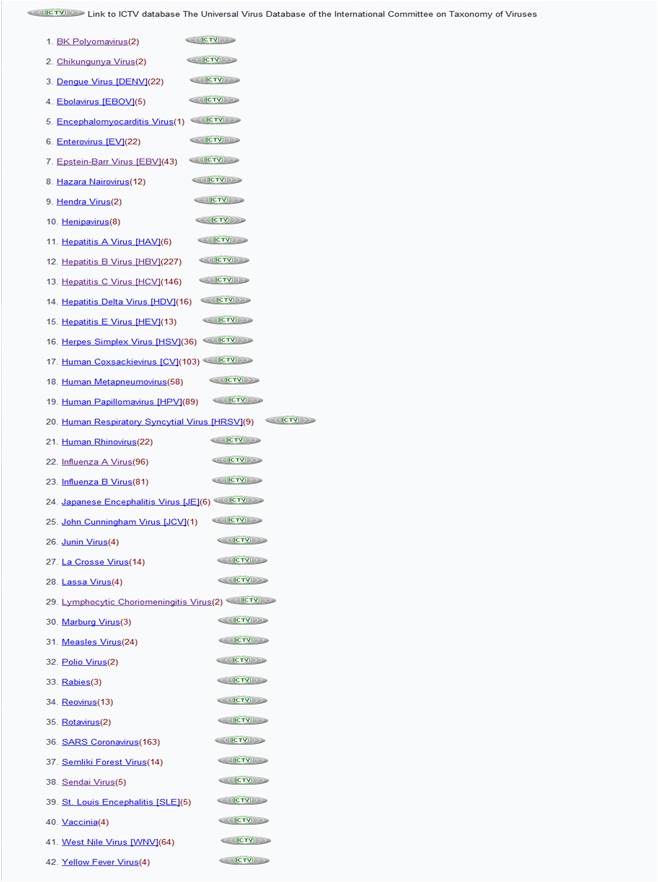
How many viruses are there in VIRsiRNAdb?
There are 42 viruse. These are:
BK polyomavirus, Chikungunya Virus, Dengue Virus [DENV], Ebolavirus [EBOV], Encephalomyocarditis virus, Enterovirus [EV], Epstein-Barr Virus [EBV], Hazara nairovirus, Hendra Virus, Henipavirus, Hepatitis A Virus [HAV], Hepatitis B Virus [HBV], Hepatitis C Virus [HCV], Hepatitis Delta virus [HDV], Hepatitis E Virus [HEV], Herpes simplex Virus [HSV], Human Coxsackievirus [CV], Human Metapneumovirus, Human Papillomavirus [HPV], Human Respiratory Syncytial Virus [HRSV], Human Rhinovirus, Influenza A Virus, Influenza B Virus, Japanese Encephalitis Virus [JE], John Cunningham virus [JCV], Junan Virus La Crosse Virus, Lassa Virus, Lymphocytic Choriomeningitis Virus Marburg Virus, Measles Virus, Polio Virus Rabies, Reovirus, Rotavirus, SARS Coronavirus, Semliki Forest Virus Sendai Virus, St. Louis Encephalitis [SLE], Vaccinia West Nile Virus [WNV], & Yellow Fever Virus.
Is this database freely available?
This open source project is freely available through this url http://crdd.osdd.net/servers/virsirnadb/
What is the use of this database?
VIRsiRNASdb will become increasingly useful for developing siRNA design tools. Additionally this database will help the user to confirm whether a given siRNA sequence or similar one has already been reported or not and also what experimental conditions were used.