Algorithm Development
Dataset
We have chosen our dataset from Viral siRNA database (VIRsiRNAdb) which contains over 1358 siRNA sequences targeting different human viruses and HIV siRNA database (HIVsir) having over 750 entries. From these databases we have selected 927 and 240 19mer sequences respectively which were having numerical (quantitative) efficacy. Further 1204 sequences from patents were also used. In addition 67 more siRNAs were collected from the literature. This combined dataset of 2294 siRNAs was reduced to 1725 sequences after removal of redundant sequences. From this dataset we randomly selected 345 sequences for validation and the rest 1380 sequences were used in training
Parameters
Various sequence features like nucleotide frequency, binary pattern, thermal stability and many hybrids approaches.
1) Nucleotide Frequency:
The objective of calculating nucleotide frequency of siRNA sequences is to transform any length of nucleotide sequence to fixed length feature vectors. This is important while using machine-learning techniques because it requires fixed length pattern. The information of each siRNA can be encapsulated to a vector of 4, 16, 64, 256 and 1024 multi-dimensions using frequency of its mononucleotide, dinucleotide, trinucleotide, tetranucleotide, pentanucleotide subsequences respectively.
2) Binary Pattern:
We employed binary pattern to extract siRNA features based on the occupancy of nucleotides at each position of siRNA sequences. Four binary patterns used for each nucleotide are follows
A=1000, C=0100, G=0010, U=0001
and this resulted in accumulation of 76 patterns for each 19-mer siRNA.
3) Thermodynamics Properties:
The thermodynamic dimensions correspond to the Gibbs free energy stability of the nucleotide pairs of the siRNAs.
4) Secondary Structure:
Secondary structure was calculated using RNAfold programme of Vienna RNA package. It predicts the minimum free energy (MFE) secondary structure and equilibrium base-pairing probabilities of single sequences.
3) Hybrids Approaches:
In hybrid approach, besides being used individually, more parameters from nucleotide frequency and binary pattern were used in order to increase the performance of the prediction method. We have used two hybrids methods, mono+di+tri+tetra+penta+binary and mono+di+tri+tetra+penta+binary+thermo which make vectors of total 1440 and 1461 features respectively.
Here is a list of siRNA features employed in building the models:
|
Model |
Features |
Pattern |
|
|
|
|||
|
1 |
Mononucleotide frequency |
4 |
|
|
2 |
Dinucleotide frequency |
16 |
|
|
3 |
Trinucleotide frequency |
64 |
|
|
4 |
Tetranucleotide frequency |
256 |
|
|
5 |
Pentanucleotide frequency |
1024 |
|
|
6 |
1+2 |
20 |
|
|
7 |
1+2+3 |
84 |
|
|
8 |
1+2+3+4 |
340 |
|
|
9 |
1+2+3+4+5 |
1364 |
|
|
10 |
Binary |
76 |
|
|
11 |
Thermodynamic features |
21 |
|
|
12 |
Secondary structure |
28 |
|
|
13 |
9+10 |
1440 |
|
|
14 |
9+10+11 |
1461 |
|
|
15 |
9+10+11+12 |
1489 |
|
Machine learning Technique
Support vector machines (SVMs) were trained with the selected sequence features to predict siRNA potency. SVM allows choosing a number of parameters and kernels. The SVMlight software package (available at http://svmlight.joachims.org/) was used to construct SVM classifiers. In this study, we used the radial basis function (RBF) kernel:
k(x ,y)=exp(-γ||x - y||2)
where x and y are two data vectors, and γ is a training parameter. In addition we also used other machine learning methods like ANN, KNN and REP Tree but the performance on SVM was best.
Evaluation
In order to evaluate performance of our models, we used Pearson’s correlation coefficient (R). All models were evaluated using ten-fold cross validation technique.
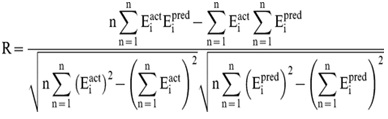
Where n is the size of test set, Eipred and Eiact is the predicted and actual efficacy respectively.