About VIRsiRNApred
VIRsiRNApred is an online webserver which predicts the numerical efficacy of siRNAs targeting different genes of human viruses. Our model is based on data collected from VIRsiRNAdb, HIVsirDB, literature and patents. The total 1725 unique siRNA sequences along with their numerical efficacy were used to build the model utilizing features like nucleotide frequency (mono to penta) , binary and thermodynamic properties. We achieved the best correlation of 0.55 PCC using hybrid features combining frequency, binary and thermo models. Our model has shown better performance compared to existing siRNA prediction algorithms, which are not trained for viral siRNAs.
How to use this server?
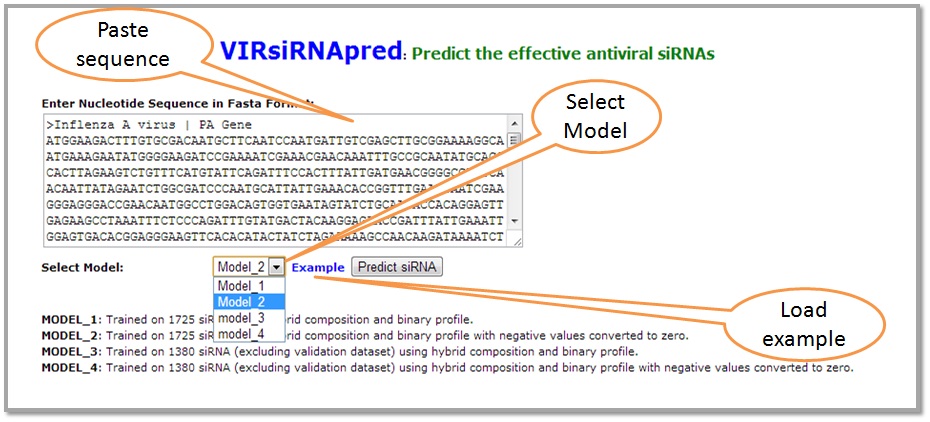
To predict the best performing siRNAs to silence a specific viral gene, simply paste the fasta sequence of the corresponding mRNA and choose the desired model and click submit.
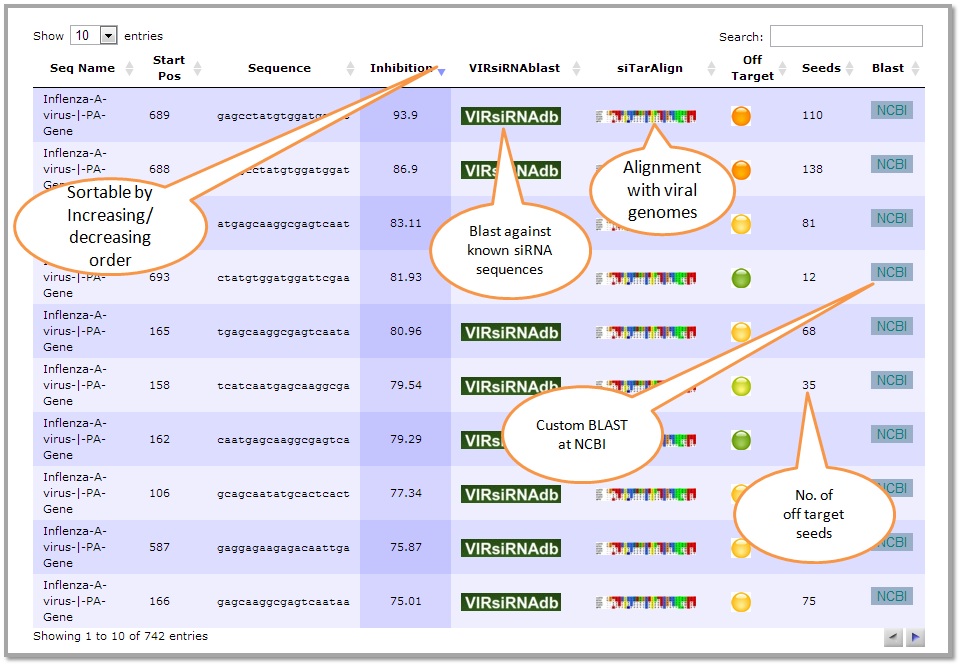
 The output table shows the recursive siRNAs chopped from the siRNA & their inhibition. Also useful links pointing to blast, alignment and off target score for each siRNA are also displayed. The user can sort the results in increasing/decreasing order.
The output table shows the recursive siRNAs chopped from the siRNA & their inhibition. Also useful links pointing to blast, alignment and off target score for each siRNA are also displayed. The user can sort the results in increasing/decreasing order.
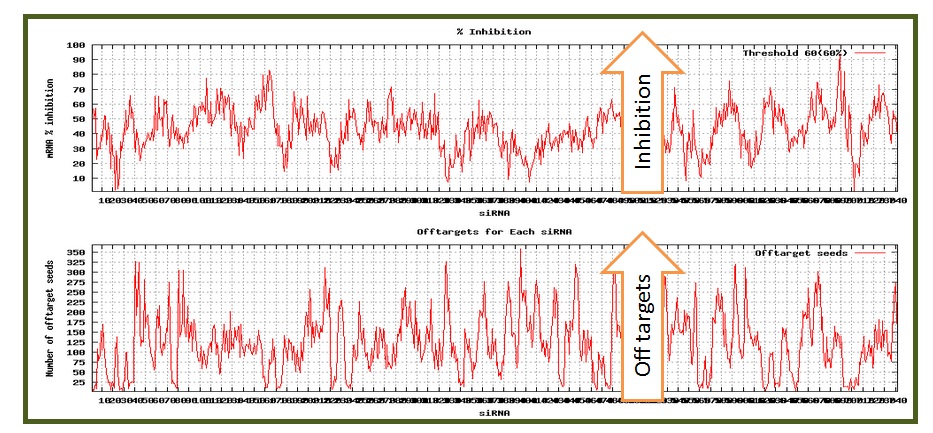
 Below the table are shown graphs depicting the inhibition and offtarget scores in a pictorial manner.
Below the table are shown graphs depicting the inhibition and offtarget scores in a pictorial manner.
 How to select the best siRNA?
1) Choose the siRNA that has the highest inhibition value by sorting the inhibition column of the result output table in decreasing order
2) Check for least number of seeds.
3) Check for minimum off targets, indicated by green (least), yellow (medium) and red (high) circles in the result output.
4) BLAST the siRNA sequence against human genome to ascertain that it does not affect other genes. Select the one with minimum matches.
5) Align the siRNA sequence with strains of viruses to check its conservation in different viral strains. Select the one which highly conserved to target maximum possible strains.
6) Lastly, you can check, whether such a siRNA has been already reported or not but using VIRsiRNAdb BLAST.
Dataset
The dataset used in this study containing 1725 siRNAs along with their corresponding id, sequence, inhibition, virus, and reference can be downloaded from the dataset page.
How to select the best siRNA?
1) Choose the siRNA that has the highest inhibition value by sorting the inhibition column of the result output table in decreasing order
2) Check for least number of seeds.
3) Check for minimum off targets, indicated by green (least), yellow (medium) and red (high) circles in the result output.
4) BLAST the siRNA sequence against human genome to ascertain that it does not affect other genes. Select the one with minimum matches.
5) Align the siRNA sequence with strains of viruses to check its conservation in different viral strains. Select the one which highly conserved to target maximum possible strains.
6) Lastly, you can check, whether such a siRNA has been already reported or not but using VIRsiRNAdb BLAST.
Dataset
The dataset used in this study containing 1725 siRNAs along with their corresponding id, sequence, inhibition, virus, and reference can be downloaded from the dataset page.