Welcome to Home Page of PCMdb |
| PCMdb is a database of methylated genes found in pancreatic cancer cell lines and tissues. In other words, it provides comprehensive information about methylation status of various genes in Pancreatic Cancer (PCa) and their comparison with transcriptomics and genomics landscape. It is a manually curated database where information is collected and compiled from literature/databases. This database will be very useful to discover novel biomakers for PCa as well as to understand the role of methylation in drug resistance.
| |
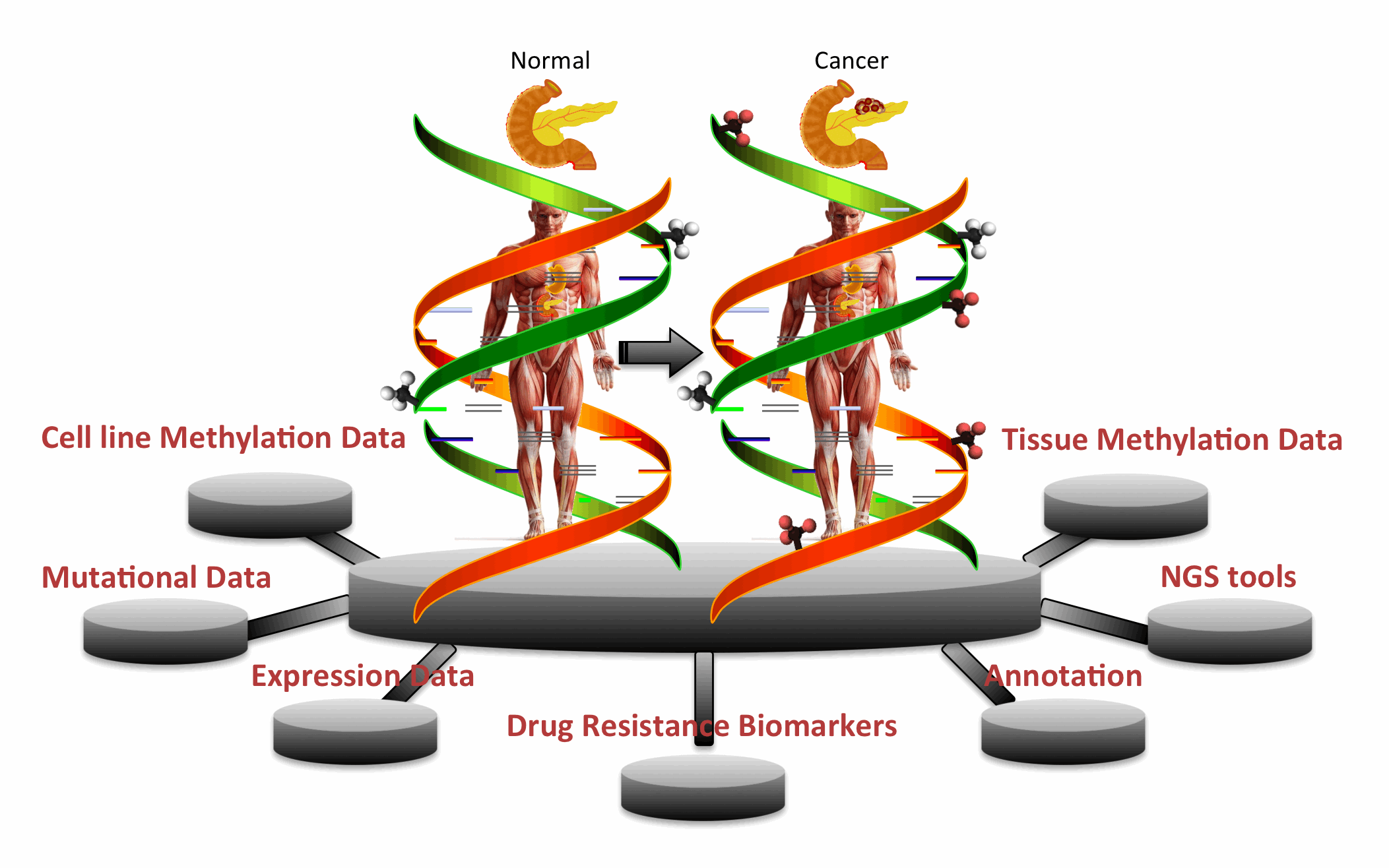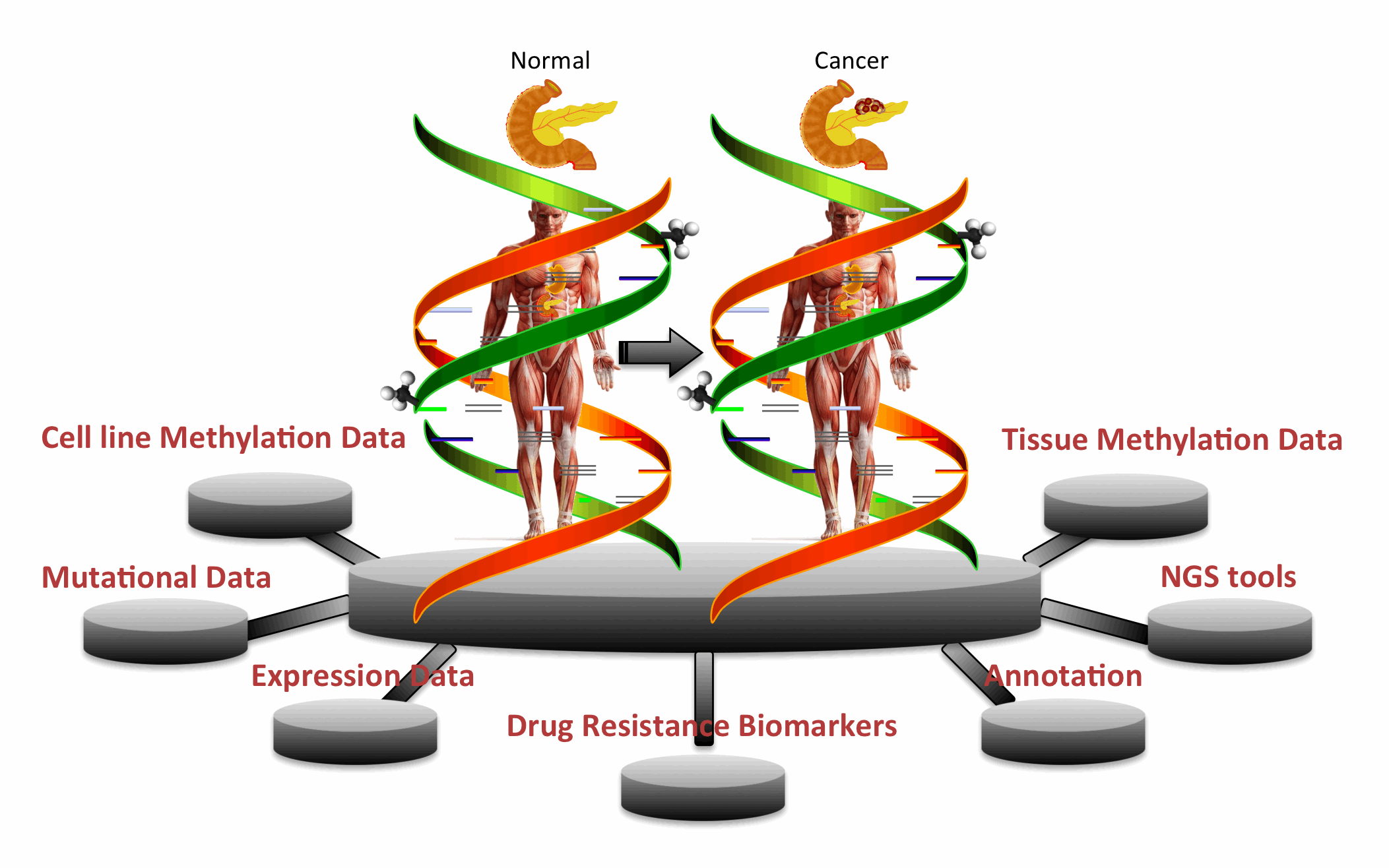  |
Major Features of PCMdbSEARCH/BROWSE: Keyword and Advanced search options allow one to retrieve data from PCMdb. Browse facility facilitates users to categorize/classify PCMdb data on different fields (Cell Lines, Genes, Chromosome, Method and Methylation Category). Integration of Tools: A large number of web based tools have been integrated into PCMdb that include similarity search techniques like BLAST and SMITH-WATERMAN. It also facilitates users to process their NGS data where it can map methylation on short Reads and Contigs. METHYLATION-DRUG SENSITIVITY: Correlation of methylation status with those of expression and CNVs and mutations is given in PCMdb which would be useful for further analysis in the field of Pancreatic Cancer treatment. UTILITY: This is a unique repository encompassing methylation status. PCMdb would be useful for finding relations of methylation of important genes with genomics and transcriptomics landscape of cells. |
| Team | Contact Us | Raghava | CSIR-IMTECH | CSIR |
