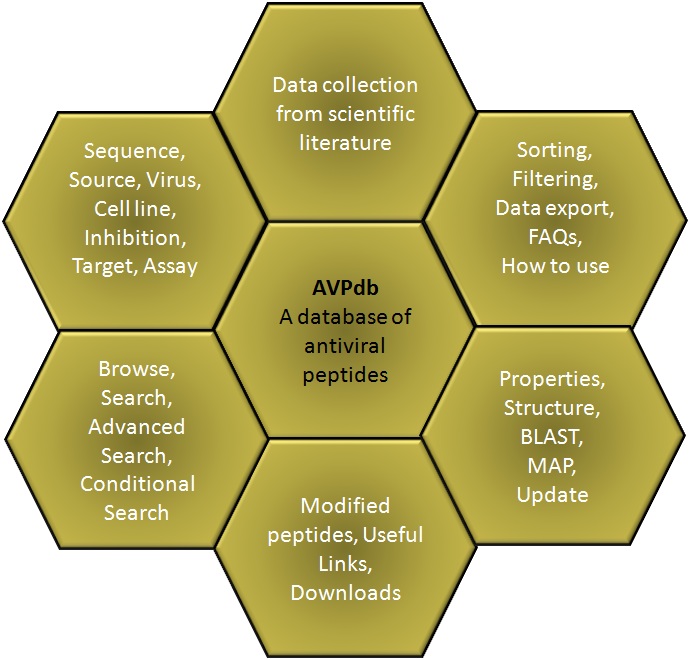
CONTENTSDatabaseBrowse Search Advanced Search Data organization Tools Update |
AVPdb is a manually curated, open source database of Antiviral Peptides (AVPs) targeted against 60 viruses. The database comprises of experimentally verified 2059 normal peptides spanned over varied efficacies and lengths, reported in the literature and tested on 85 cell lines. 624 modified peptides having additional chemical groups are presented under Modified AVPs. Database also provides the user with physicochemical properties and structure of AVPs along with more informative tools for data analysis such as BLAST and MAP as well as links to major peptide resources. The database comes with enhanced user experience and easy to operate browsing as well as searching with sorting and filtering functionalities. This database has been dedicatedly designed, referring to the current needs of scientific community working exhaustively for antiviral therapeutic development and is anticipated to be useful for them who are seeking a committed repository for AVPs. The database is freely available at http://crdd.osdd.net/servers/avpdb.
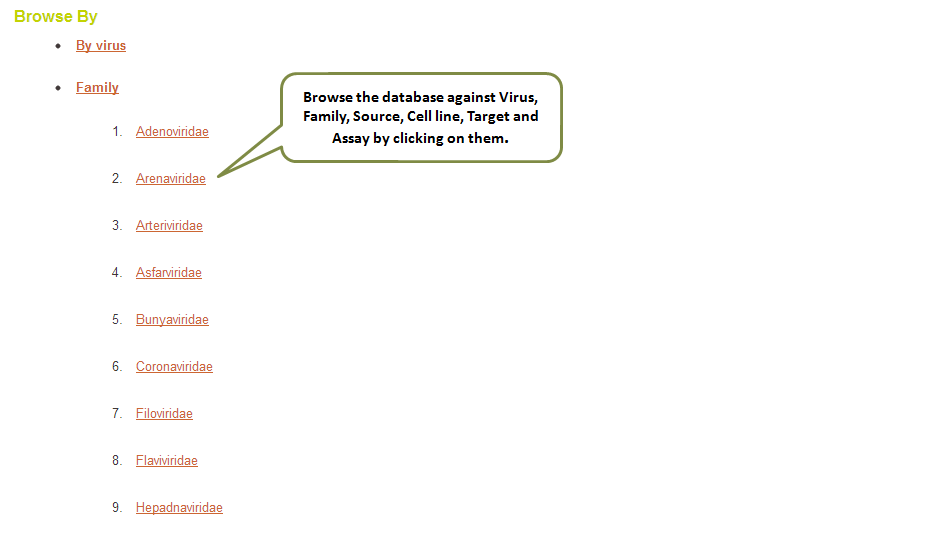
This user-friendly “Browse by” option allows you to explore the data for normal peptides by any of the fields categorized in the database viz. Virus, Family, Peptide Source, Cell Line, Target and Assay. For modified AVPs also, a separate browse option is provided where the data can be sought by Virus, Modification, Peptide Source, Cell Line, Target and Assay.
The database has been incorporated with four different searches:
1) Field Search:
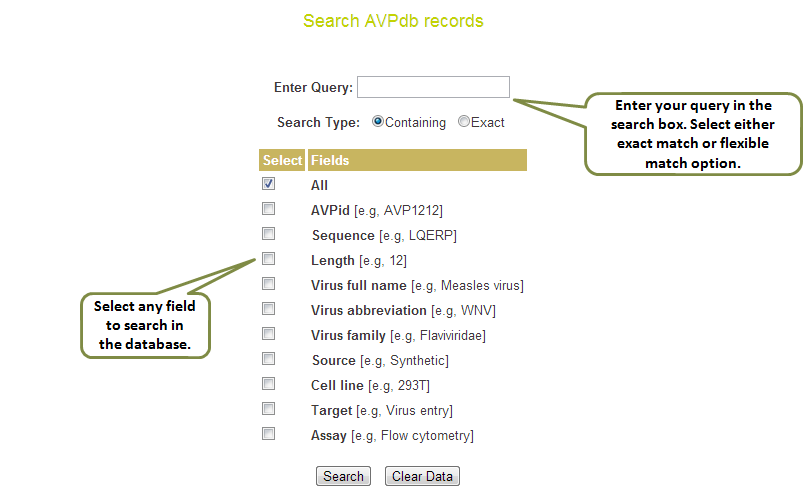
Input: Here you can enter your query in the box and can specify any of the 10 fields against which you wish to search or else you can keep the default “All” option which will search against all the fields in the database. Examples have been provided corresponding to each field which can be used as sample queries. Besides the option to choose the field, search type allows to retrieve either an exact match or the match containing the query. Clicking on clear data button clears the data after your search.
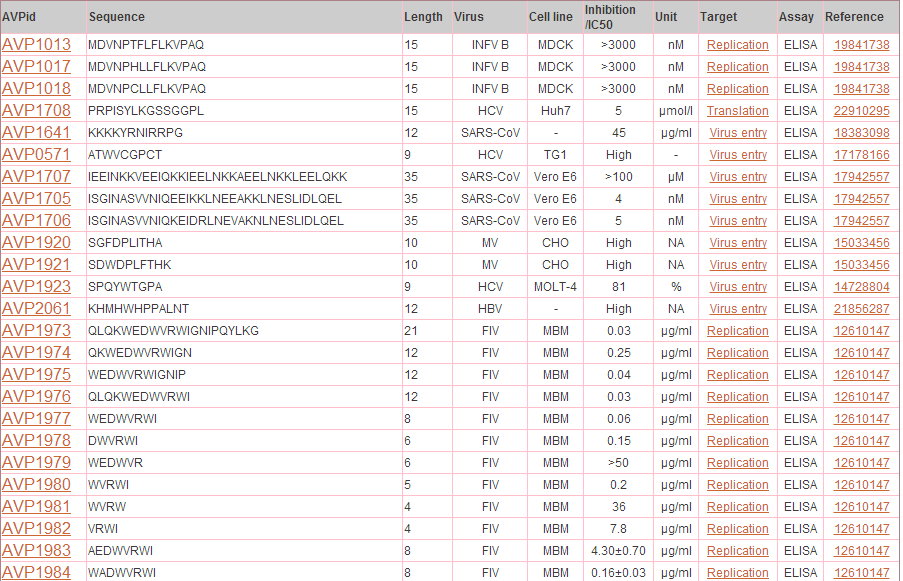
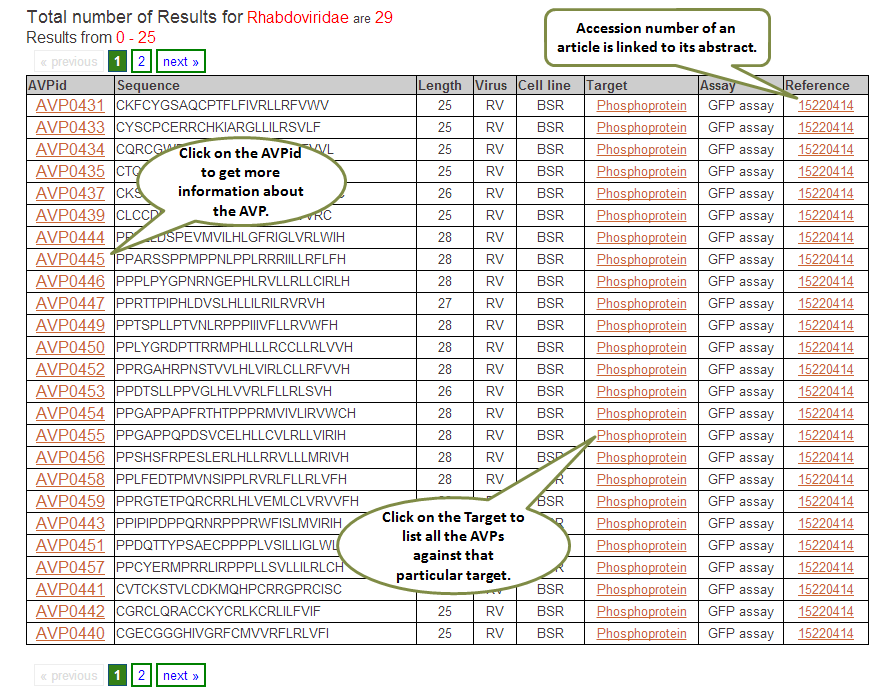
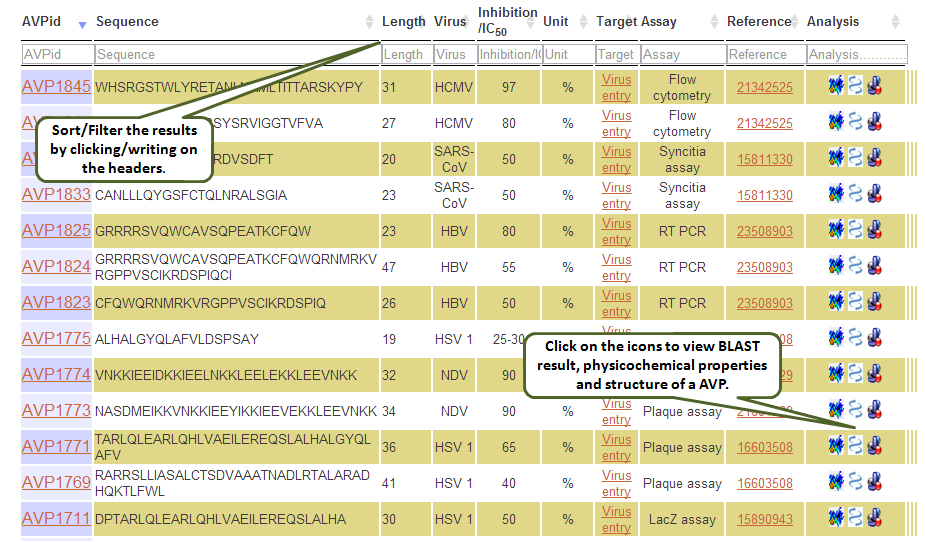
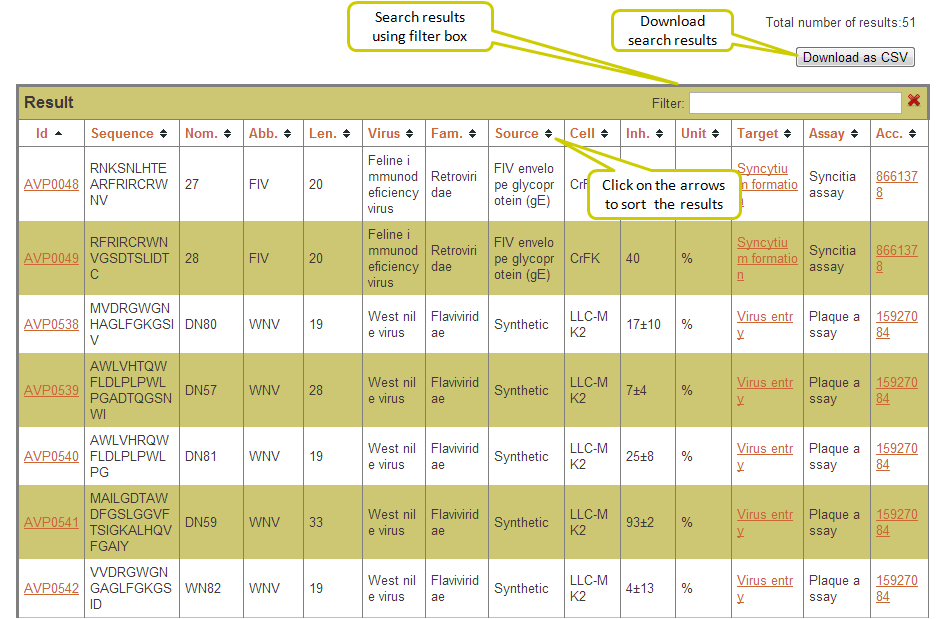
Output: The results obtained from this search display 10 fields where first 9 contain the experimental data and the last one, Analysis, has links to BLAST results, physicochemical properties and predicted peptide structure. The entries in brick-red colour are further linked. For example, when you click say, Virus entry under Target column, all the peptides in the database having Virus entry as target are displayed. The entries under Reference column redirects you to the full article in Pubmed. The results retrieved here can be exported to a comma separated value (.csv) file. You can sort the data by clicking on the column header and filtering can be done by typing in the boxes given under the headers.
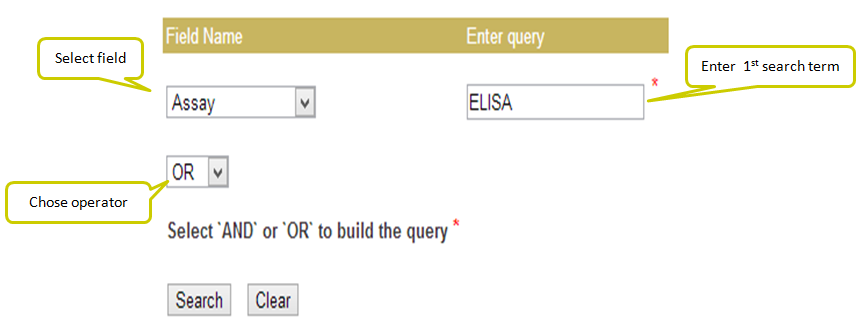
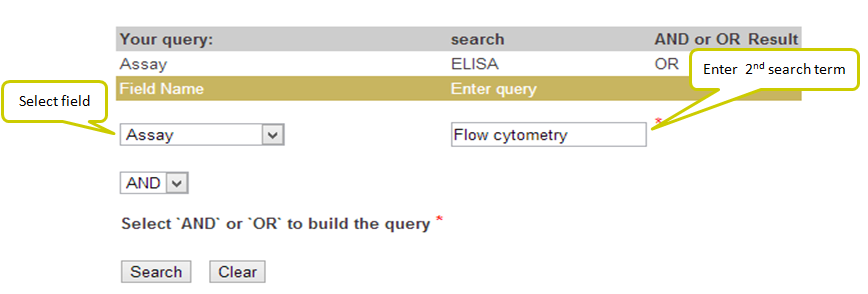
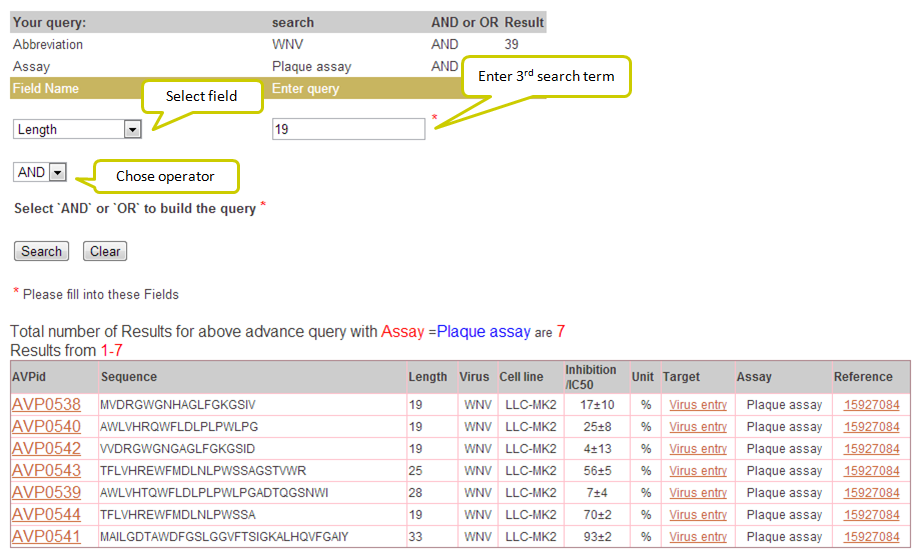
Input: This feature helps you to search this database using logical operators (AND/OR). For example, you have obtained say 39 results for WNV against Virus abbreviation field but you wish to filter those AVPs with assay as Plaque assay and of length 19. For such results, you can use AND operator alongwith choosing the desired fields.
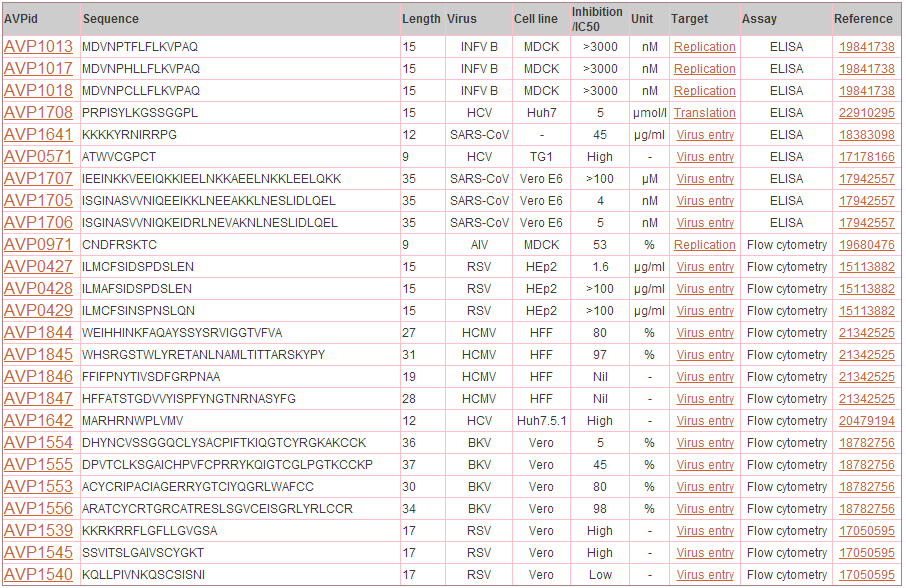
Output: In the example below results obtained from this search will retrieve the AVPs corresponding to Virus abbreviation = WNV and Assay = Plaque assay and Peptide length = 19.




To view another example [+] Click here.
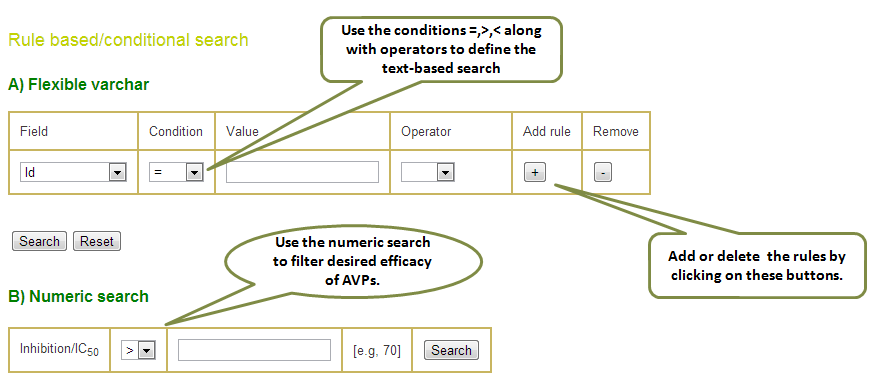
3) Conditional Search
Input: This additional search facility allows you to add conditions like >,<,= or LIKE to define your query and also helps building the searches using logical operators in a single query. This search has further two divisions a) Flexible varchar (variable character) search and b) Numerical search. The former is suitable for variable text based search while the latter is optimised for numbers.
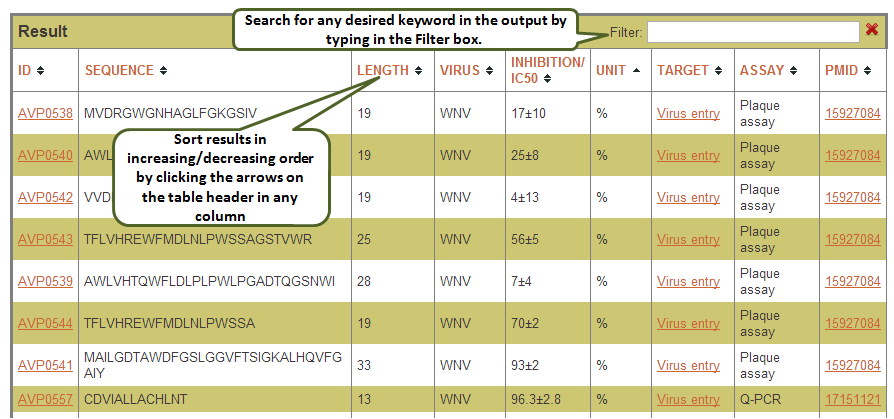
Output: The result table can be sorted in increasing/decreasing order by clicking the arrows on the table header in any column. Also, one can search for any desired keyword in the output by typing the word in the Filter box.
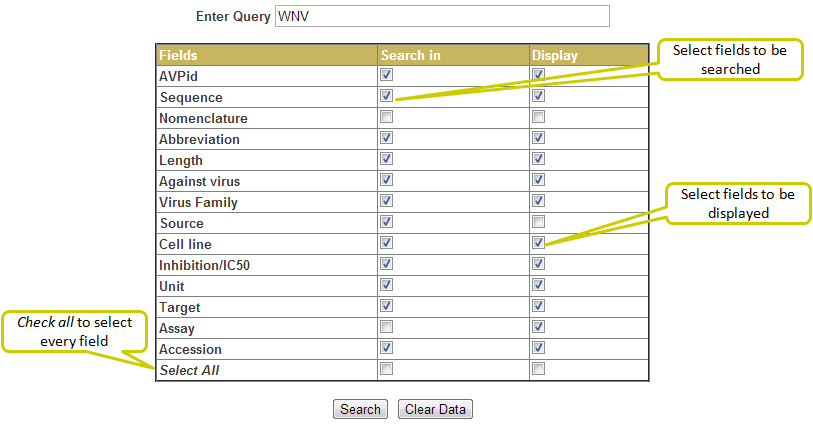
Here you can select the fields to be searched as well as the fields to be displayed (using checkboxes) in a flexible manner. This search enables the user to display multiple fields and records. The result output can be further filtered using the filter box provided at top. Also the user can download the search results (as csv file) by clicking on the download button.
Data organization
The data is organized as displayed below. After clicking on the AVPid say, AVP1709, you will be redirected to all the details corresponding to this entry: Sequence, nomenclature, length, against virus, family, source, cell line, inhibition (IC50/EC50/IC90/Ki/Kd/%/High/Low etc), unit, target, assay, properties, structure, paper, author, reference and accession number. Here also, the entries in brick-red colour are further linked.
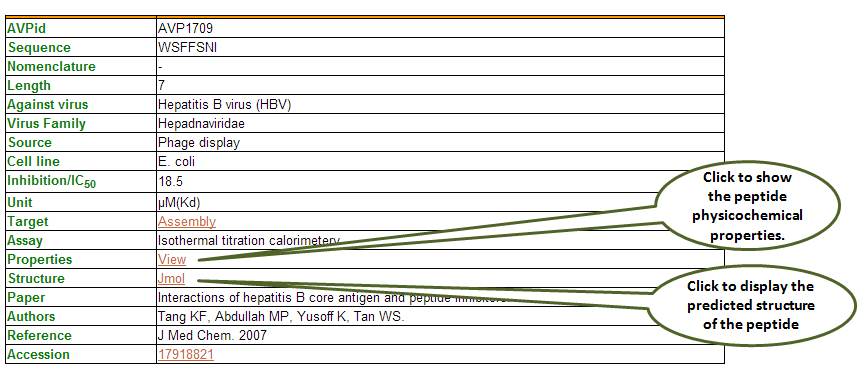
Physicochemical properties: Various important physicochemical properties such as Amino acid composition, hydrophobicity, preference for beta sheets, frequency of alpha helix, amino acid charge and polarity have been calculated using AAindex (Kawashima et al. 2008) and are included for each entry in the database. These properties can be viewed by either clicking on the physicochemical properties icon under the analysis column of field search output or by clinking on the View properties corresponding to each AVPid entry on the individual record page.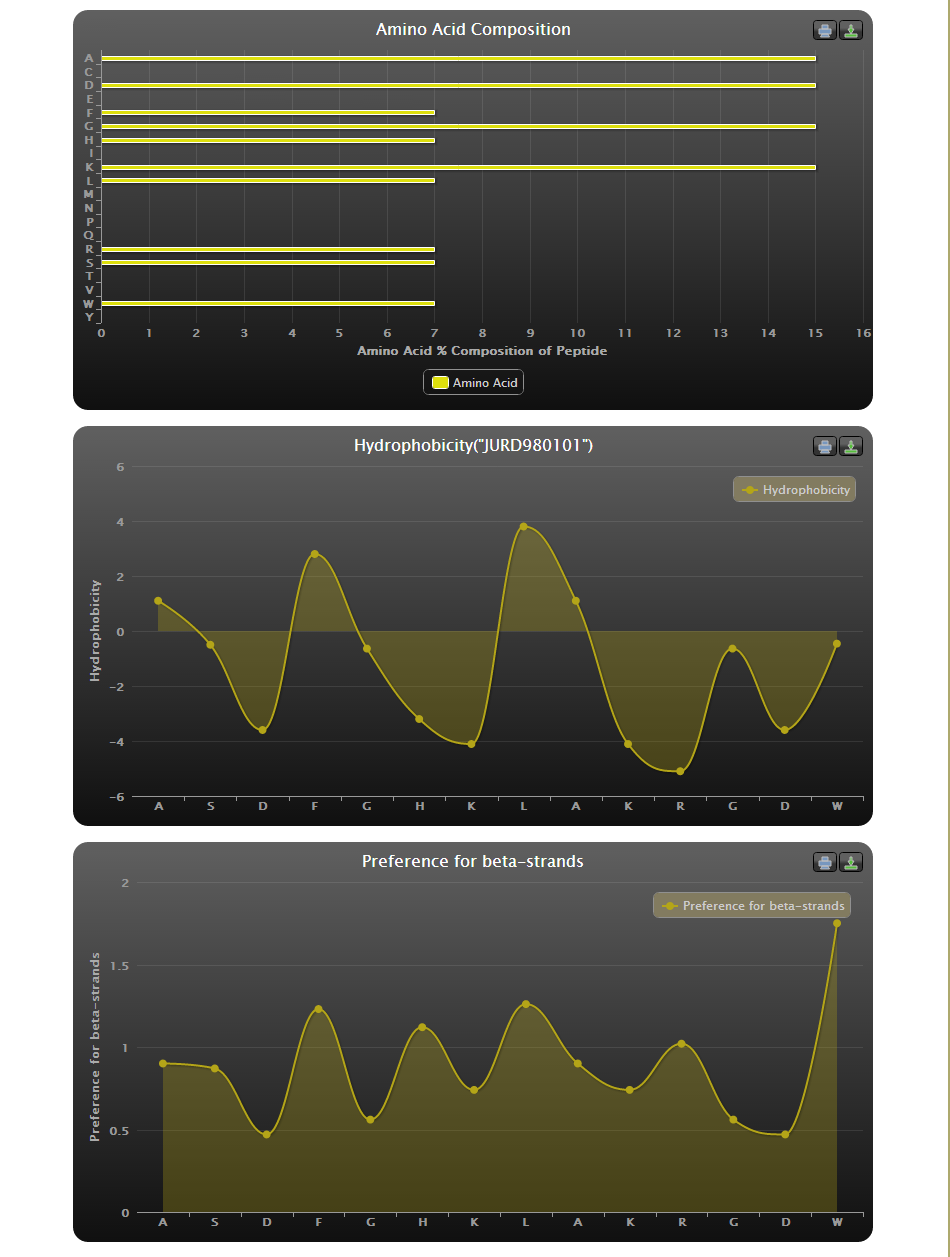
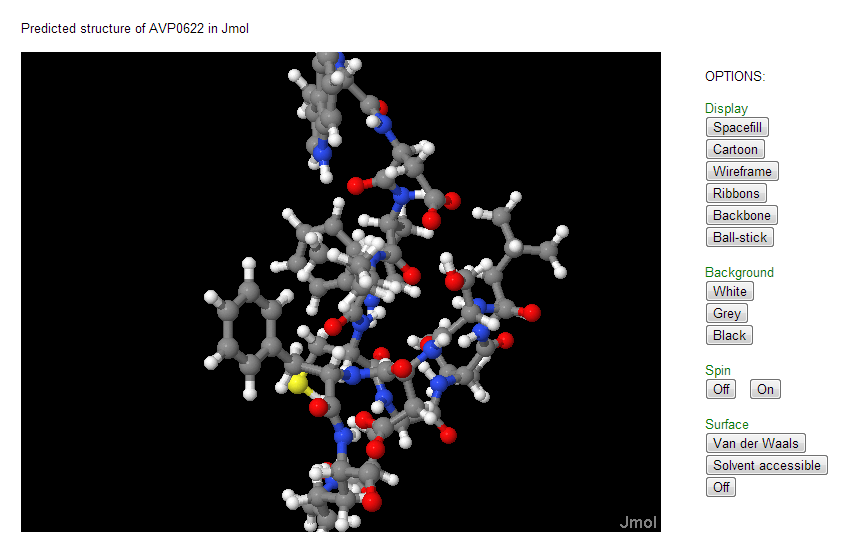
Structure: Predicted structures for all the AVPs can be viewed using Jmol which has been incorporated under the analysis column of field search as well as on the details page of each AVP. Structures are predicted using PEPstr method (Kaur et al. 2007) and PEP-FOLD server (Thevenet et al. 2012).
Note: To display the structures in your browser, please ensure you have an updated version of JavaTM installed on your system and the JavaTM plugin should also be enabled in the browser.
AVPdb Map
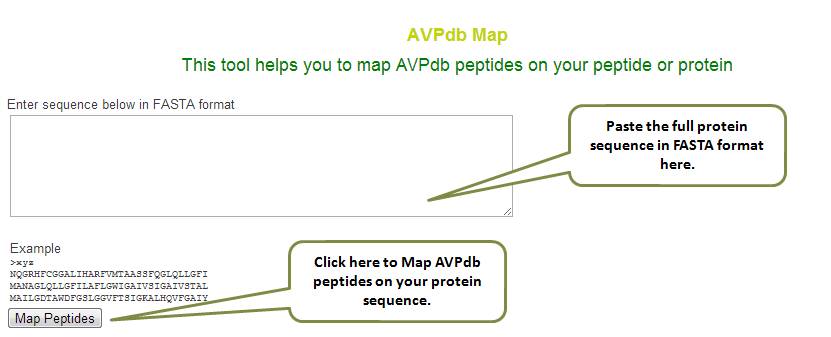
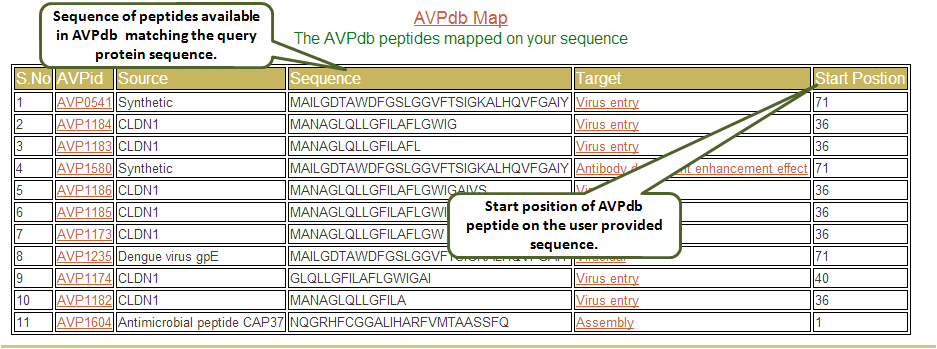
The ‘AVPdb Map’ is a user friendly tool to fetch the perfectly matching peptide available in our database. So, it helps the user to find, how many peptides against the user provided protein sequence are available in our database.
The output of this tool displays the AVPid, its source, sequence, and its target. Also mentioned is the start position where the match is found in the user provided sequence.
AVPdb Blast
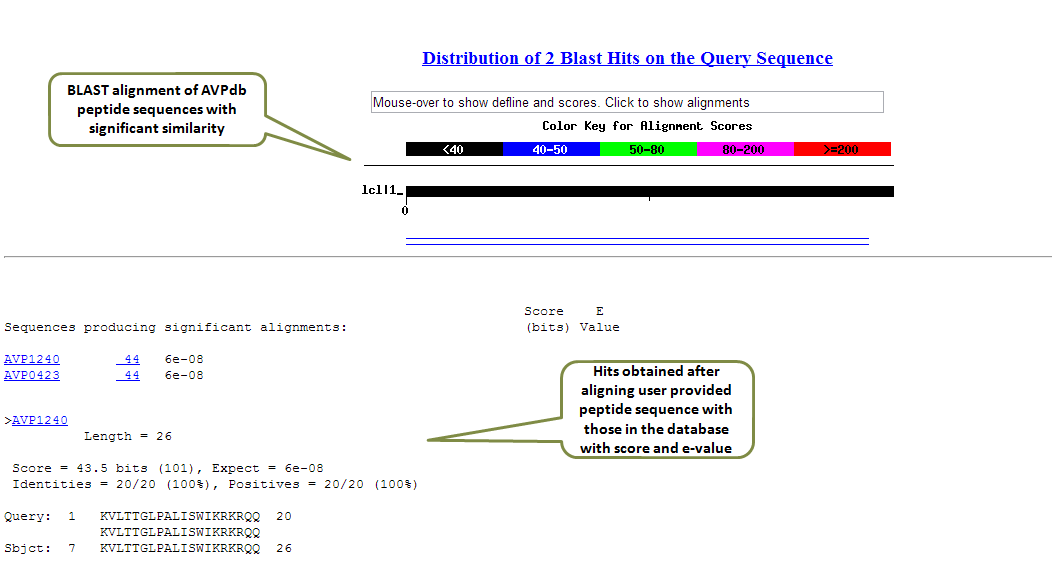
Additionally, the Blast allows alignment of a user provided peptide sequence against all the peptide sequences available in our database. This helps the user to confirm whether a given peptide sequence or similar one has already been reported or not.
The output is given in the standard format with the blast score and e-value. The alignment is shown for the peptides found to be identical or similar in the database. The output can be formatted based on the options provided by user.

Authors generating experimental data on anti-viral peptides are encouraged to submit the data directly into AVPdb. For this purpose, a web form for data submission is provided. Submitted information will be included in the database update after ascertaining its authenticity.