siRNAmod database
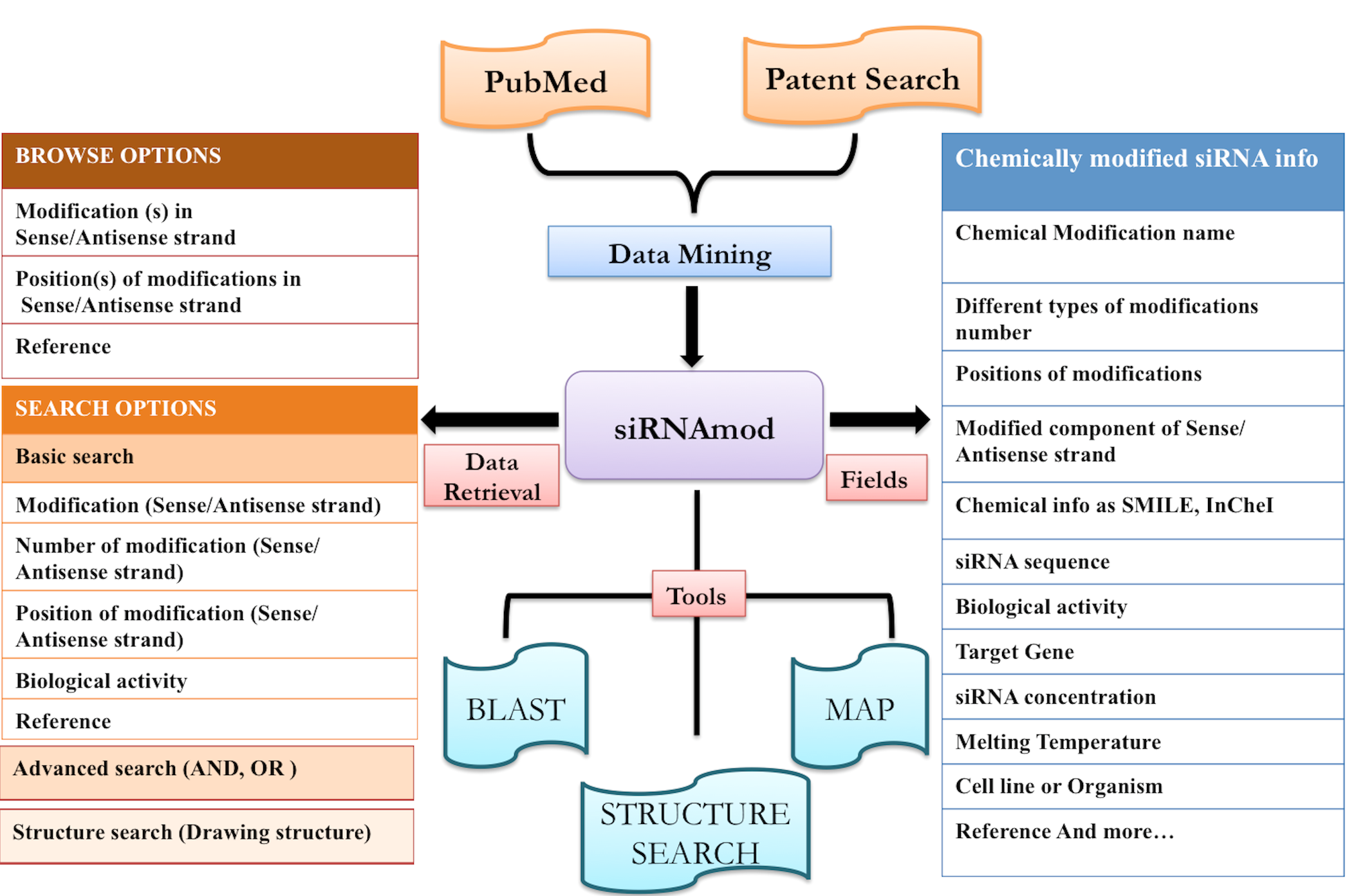
siRNAmod is a manually curated database for experimentally validated chemically modified siRNA involving 125 different types chemical modifications with different permutations and combinations of targeting various genes with 4894 siRNA sequences that are involved in cellular processes. siRNAmod will be useful for people working on siRNA-based techniques as well as chemists working in the field of improving the siRNA-based techniques. The database also provides informative tools likestructure search, BLAST and MAP as well as links to other important siRNA databases. This database is freely available through this url: http://crdd.osdd.net/servers/sirnamod/
Browse
User-friendly link allows to explore the database by any of the given fields categorized in the database as follows:
Modification in sense/antisense strand, Modification position in sense/antisense strand and Reference.

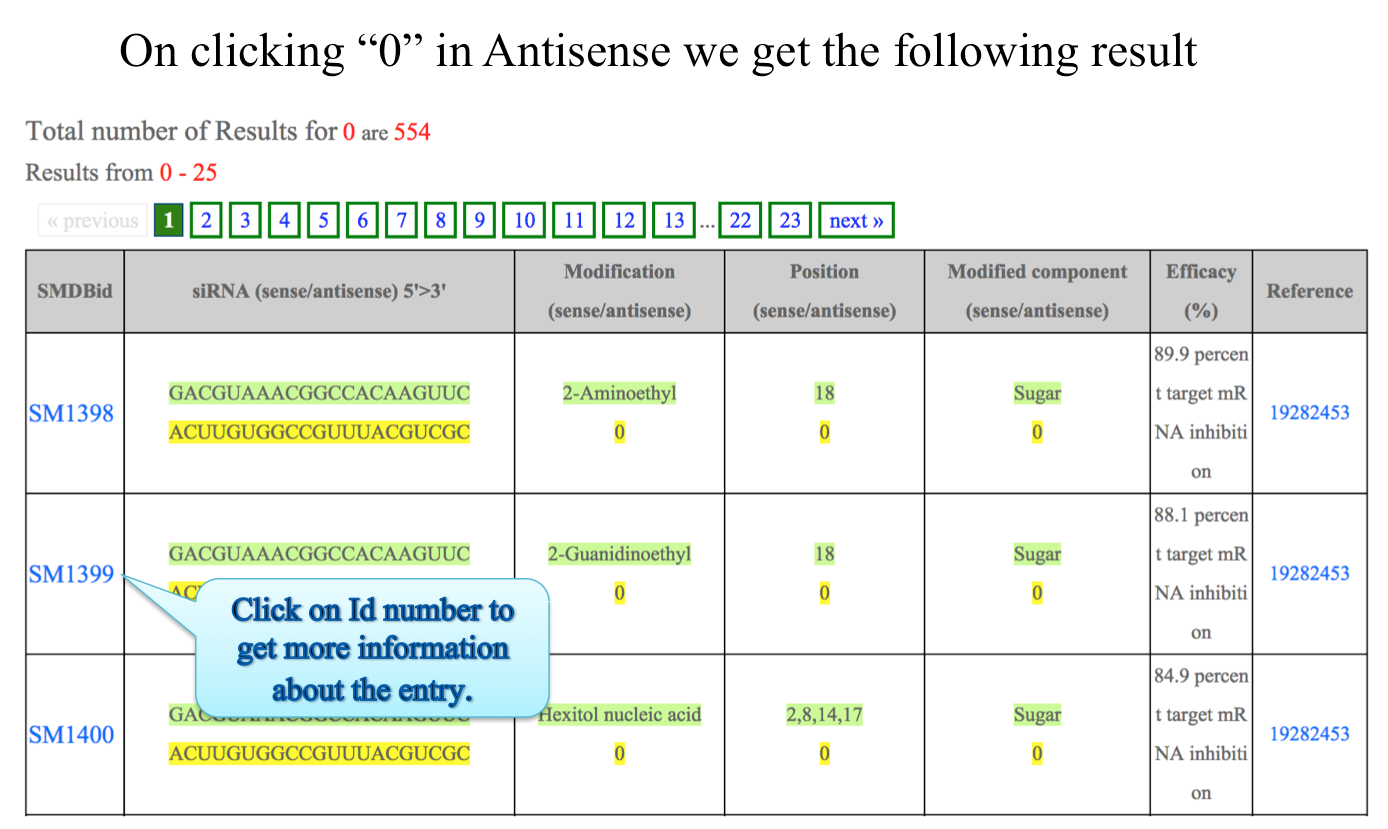
Individual entry details
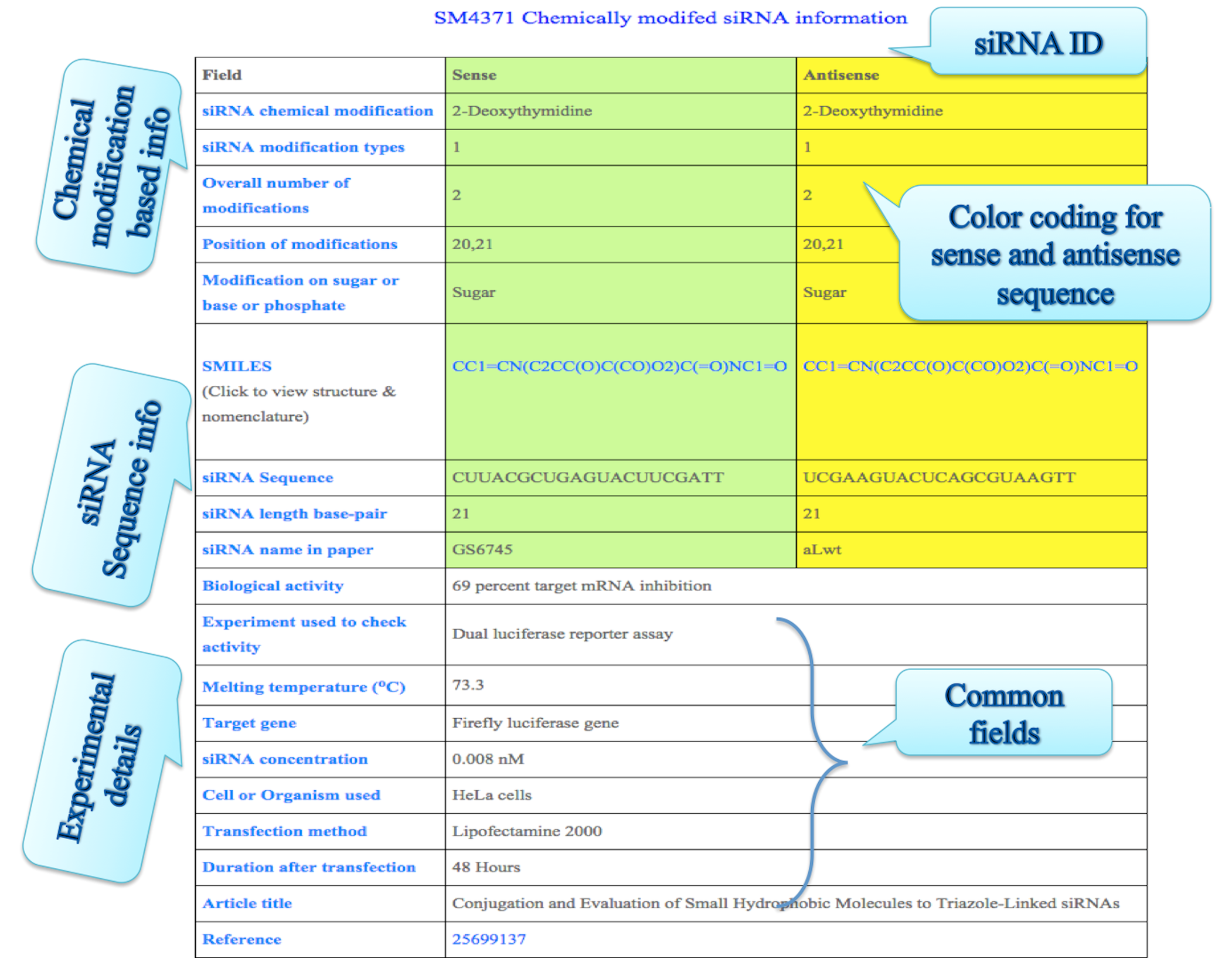
On clicking the SMILE link above we get the structure of the indivudual modification along with other information.

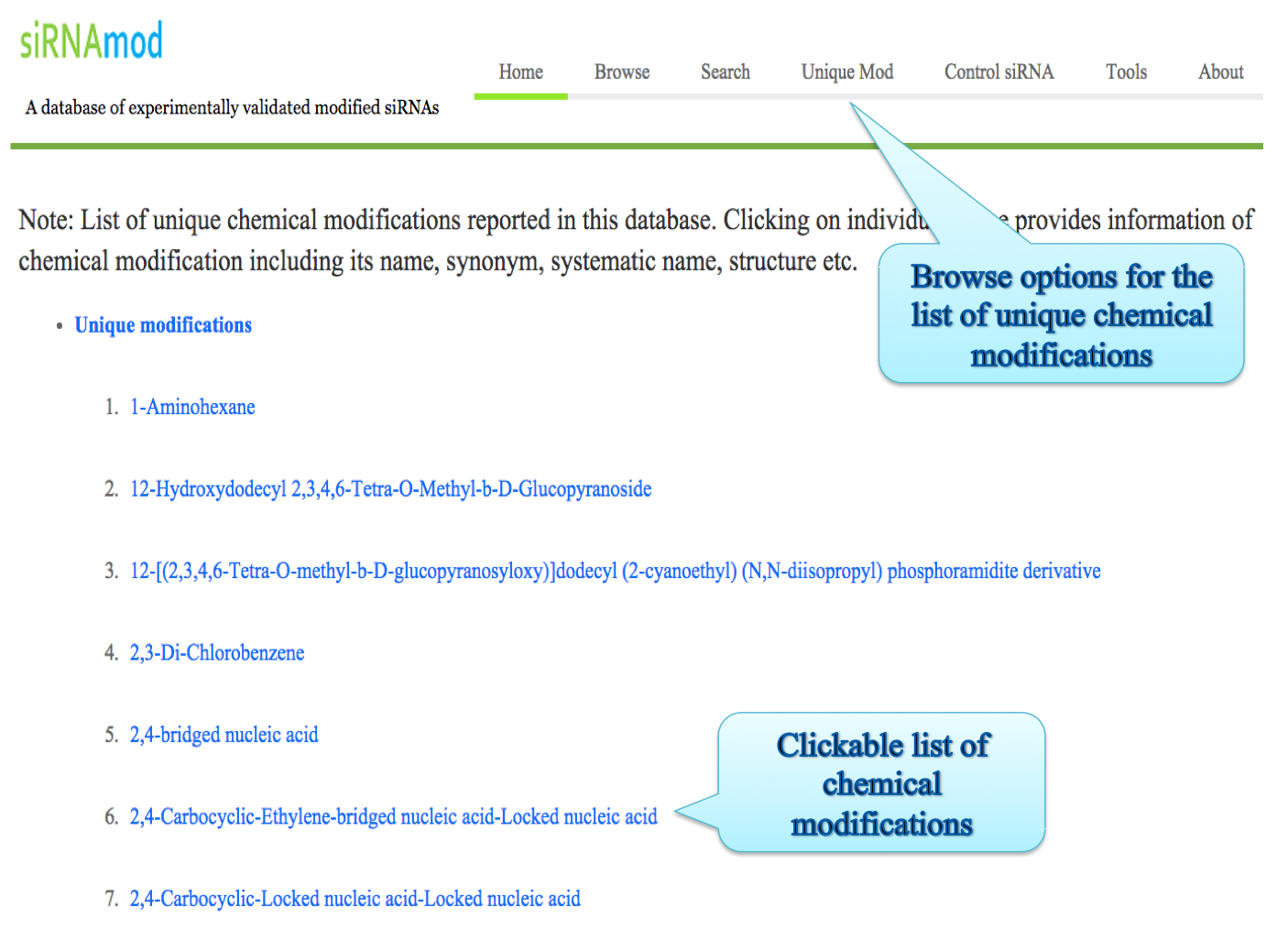
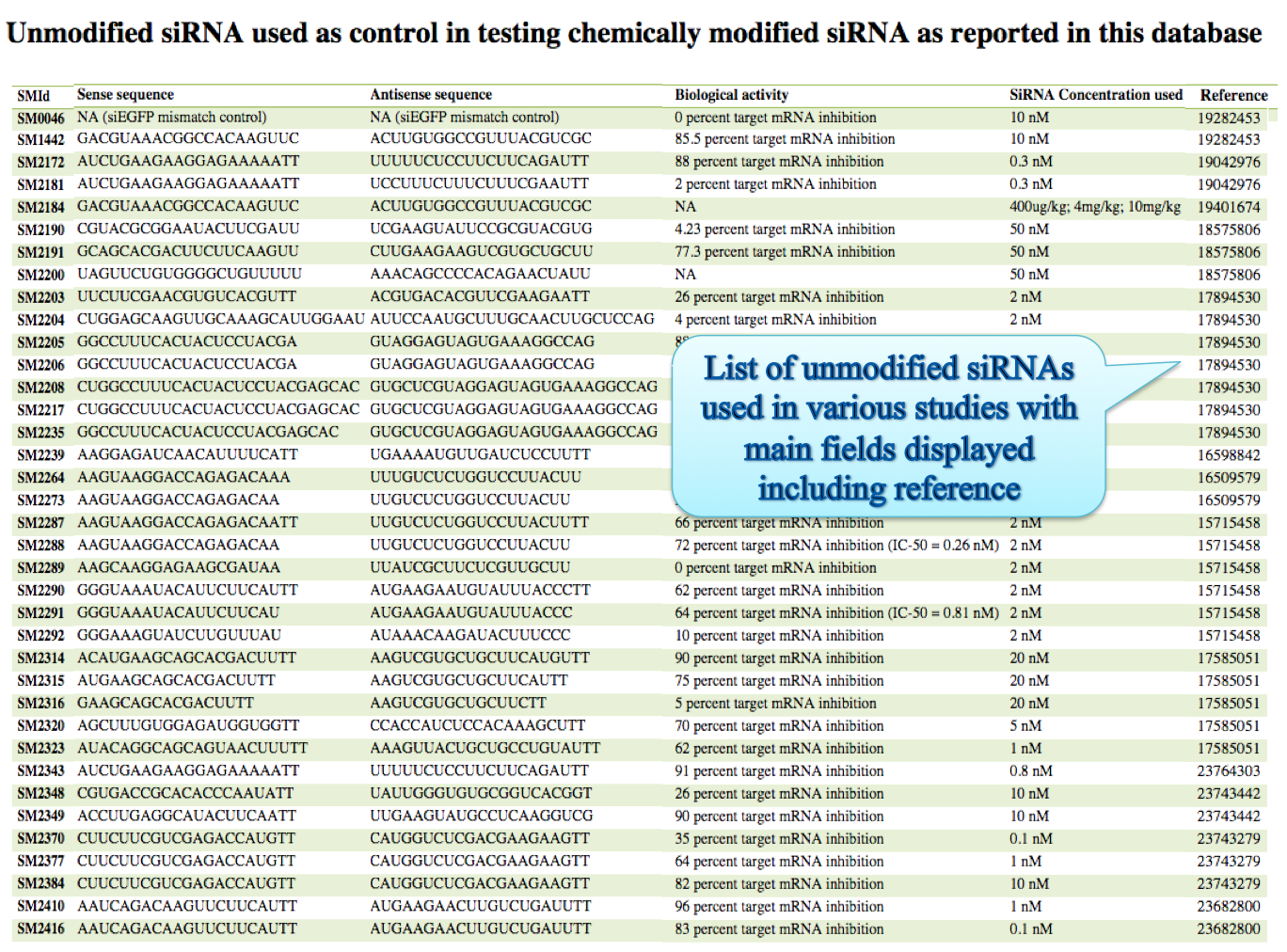
(ii) Besides unique chemical modification list is also provided. User can find all the list of modifcations here and proceed. Control siRNAs used in different studies is also provided in such manner in another browse page.
Search
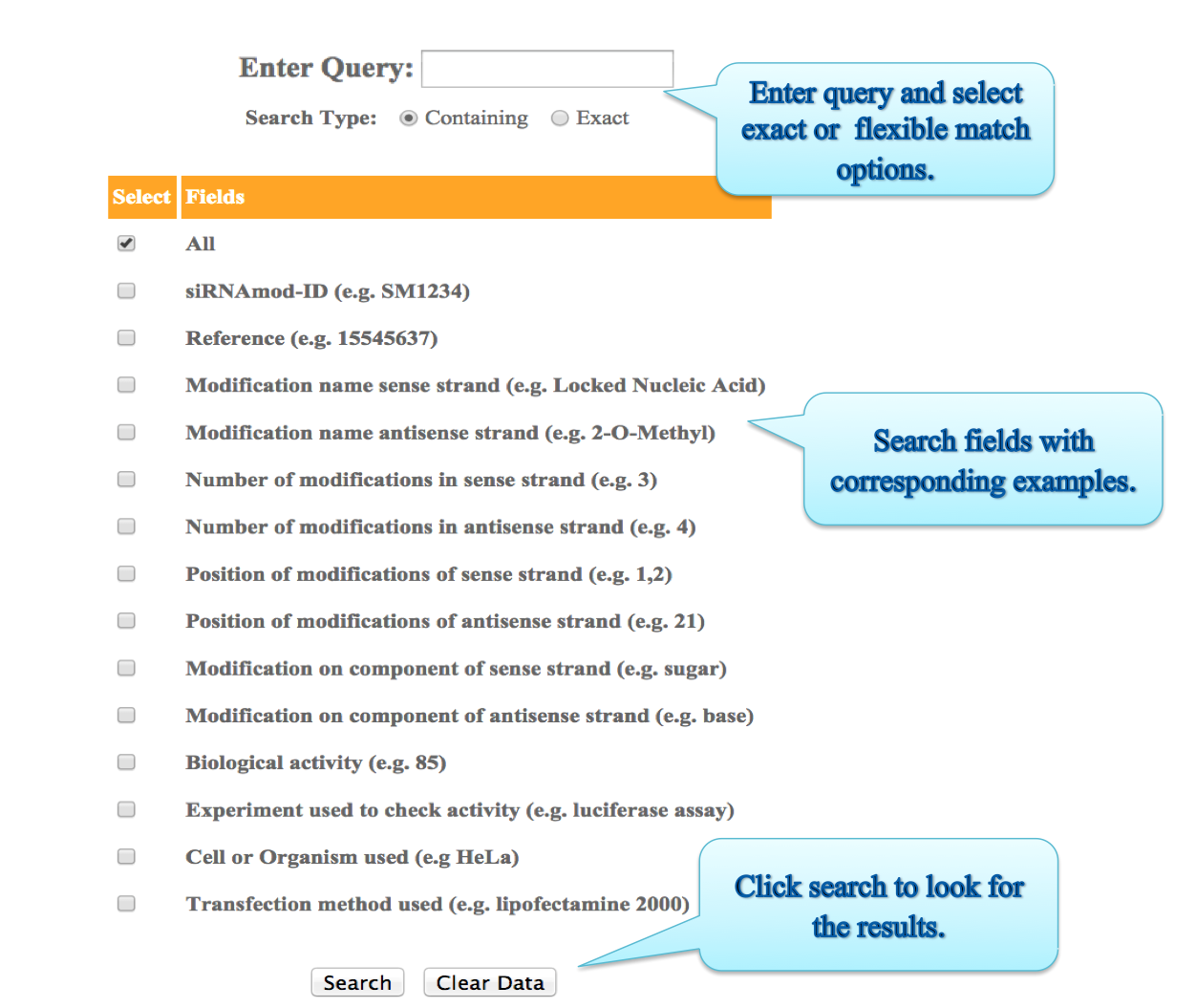
Here user can search in two ways:
i) Basic Search:
Input: Here you can enter your query in the box and can specify any of the given fields like siRNAmodID, reference (PMID),Modification (sense strand), Number of modifications in sense strand, Position of modifications of sense strand, Modification on component of sense strand,Biological activity, Modification name (antisense strand), Number of modifications in antisense strand, Position of modifications of antisense strand, Modification on component of antisense strand, siRNA concentration used,Experiment used to check activity,Transfection method used or else you can keep the default as All option which will search against all the fields in the database. Examples have been provided corresponding to each field, which can be used as sample queries. Besides the option to choose the field, search type allows to retrieve either an exact match or the match containing the query. Clicking on clear data button clears the data after your search.
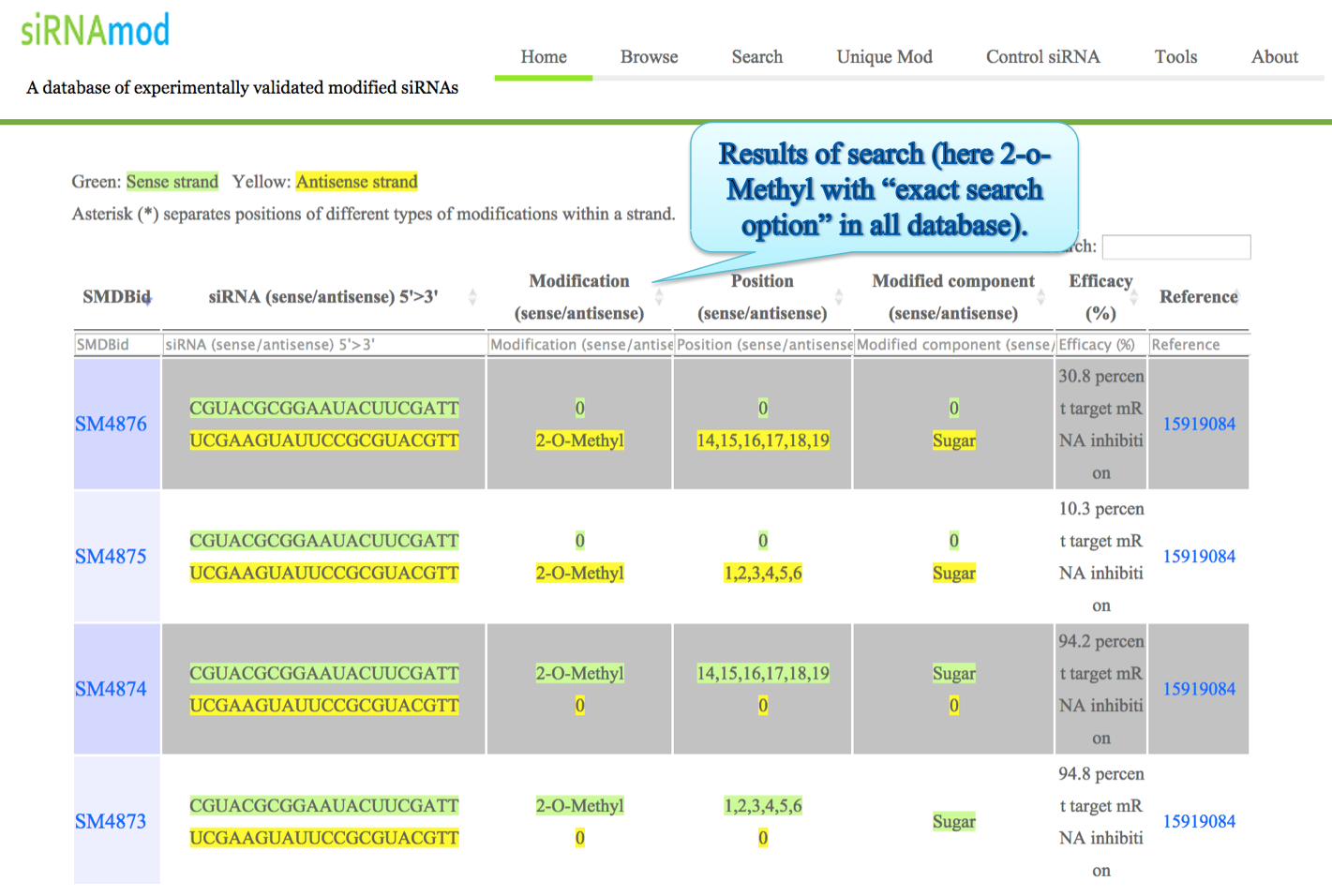
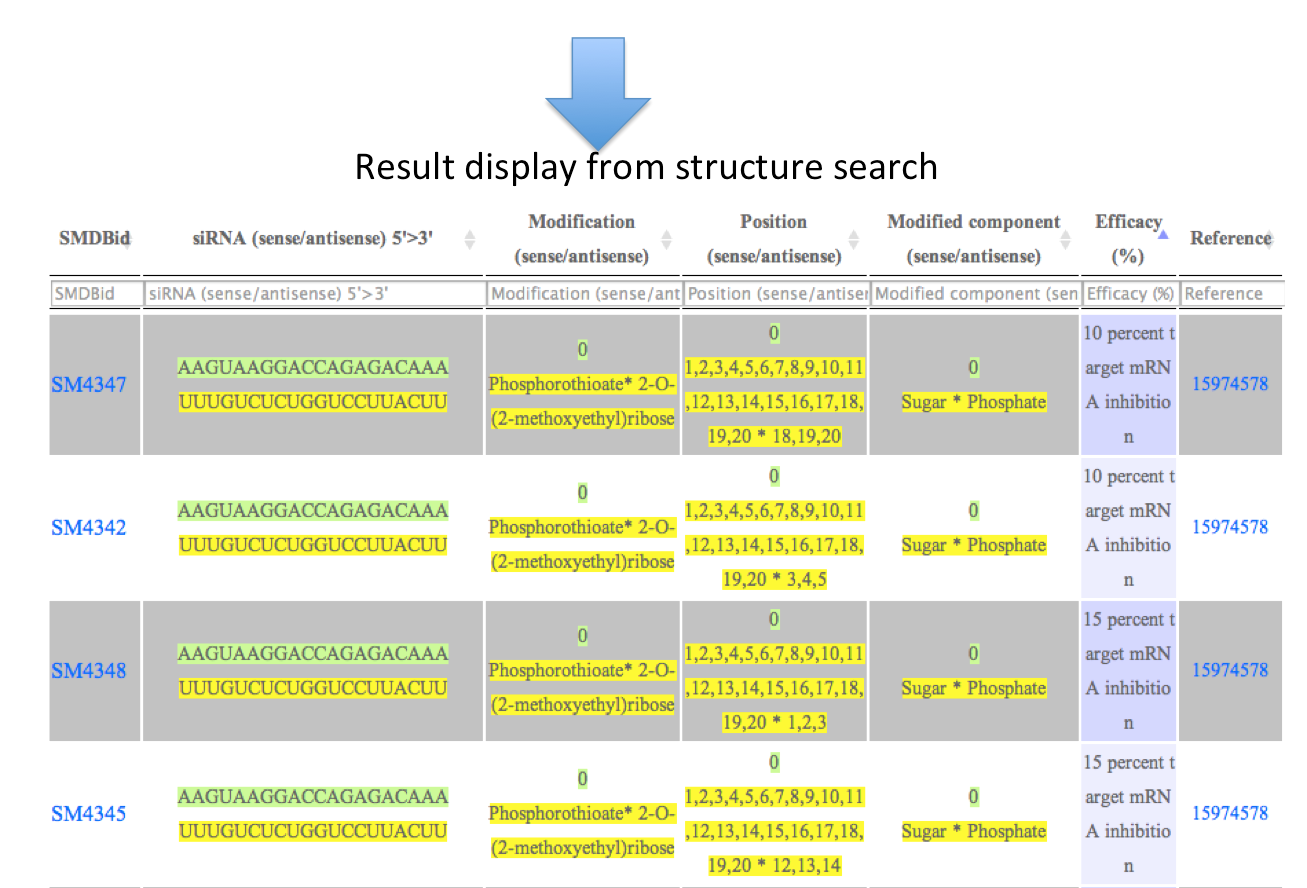
Output: The results obtained from this search display 7 fields. You can sort the data by clicking on the column header and typing in the boxes given under the headers can do filtering.
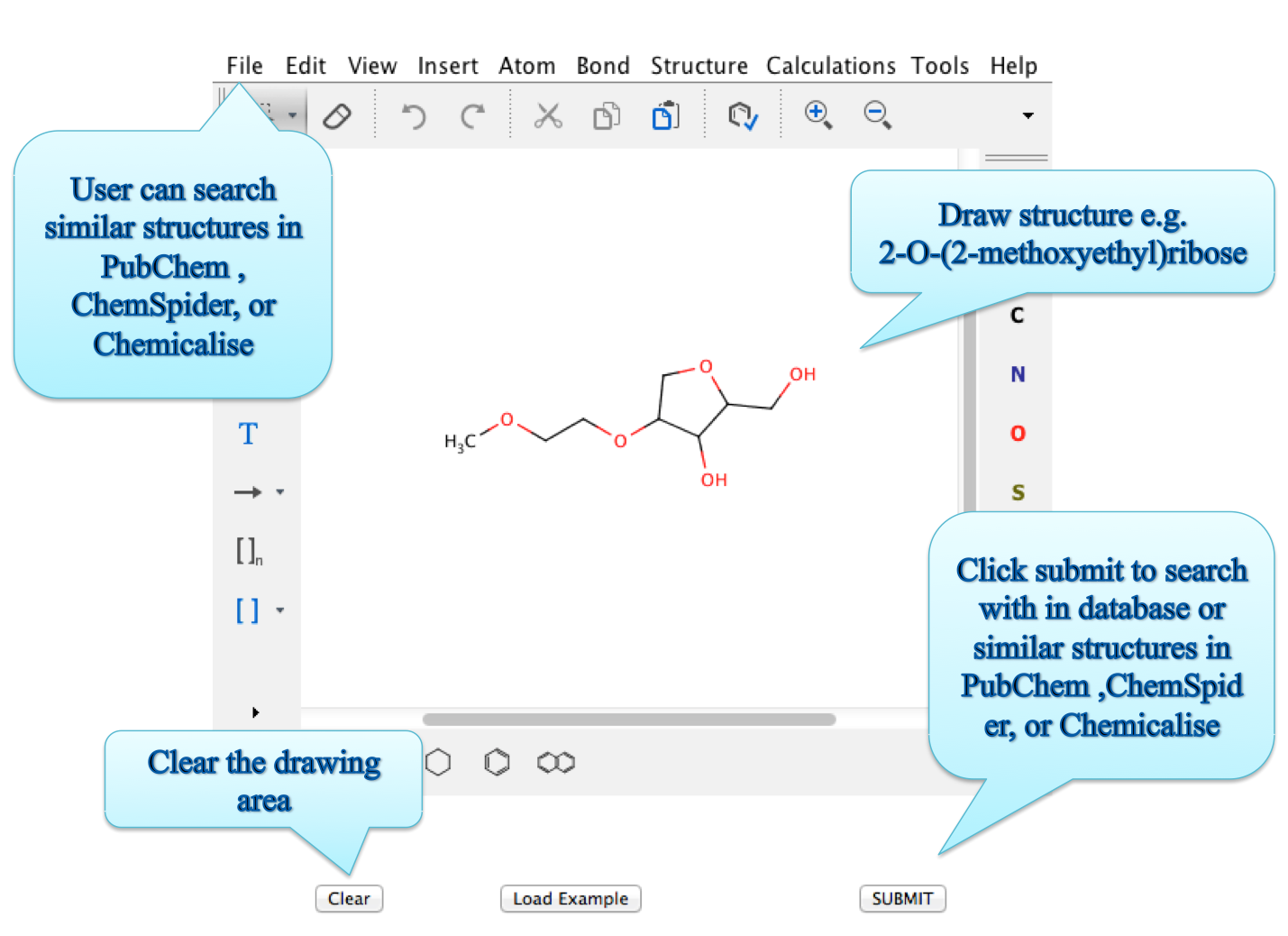
ii) Structure search:
Create the structure of the chemical and check wheather it is present in the database or not. User can even look for external databases as defined in the picture below.

Tools
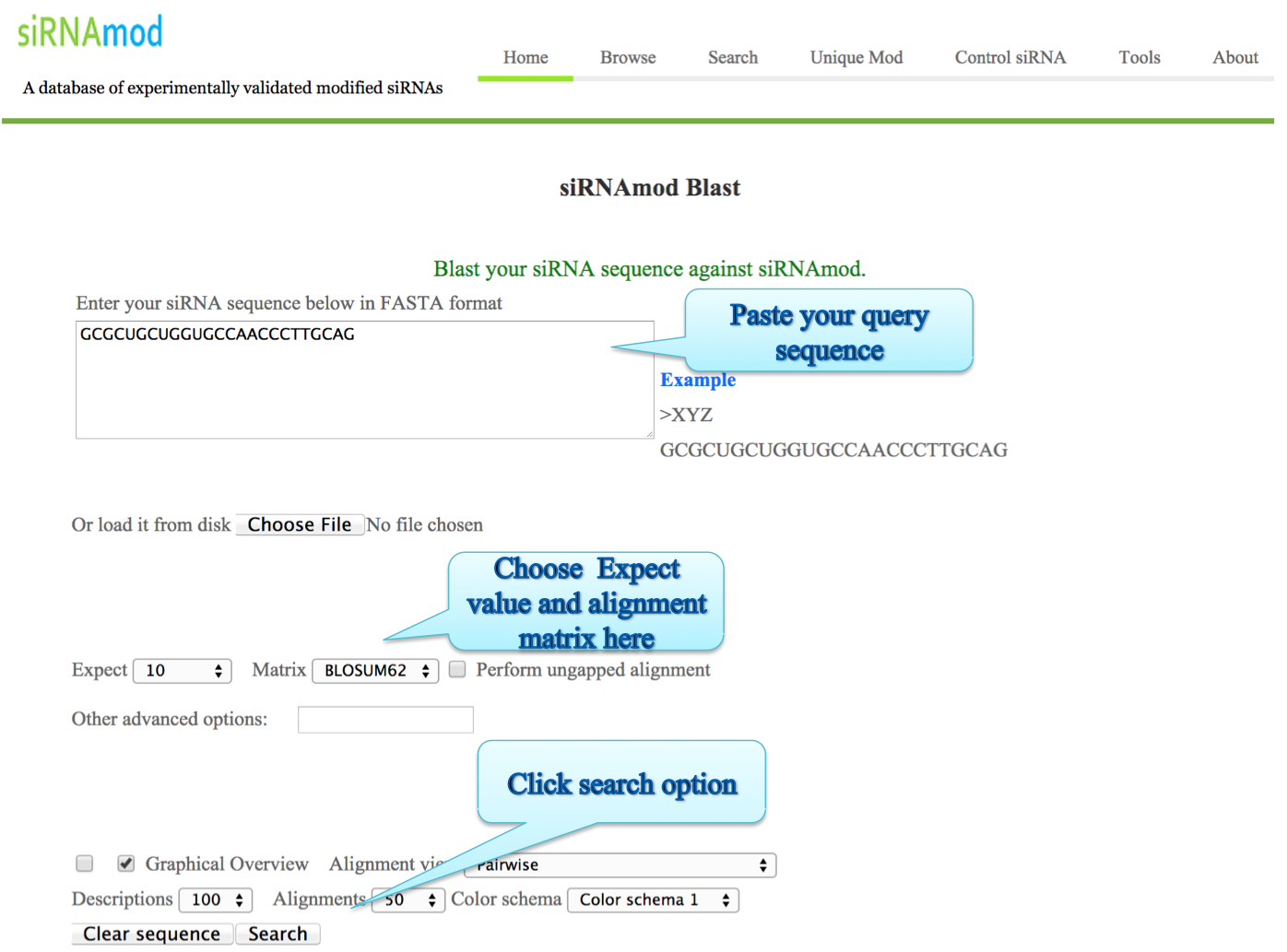
i) BLAST:
The BLAST allows alignment of a user provided siRNA sequence against all the sequences available in our database. This helps the user to confirm whether a given sequence or similar one has already been reported or not.

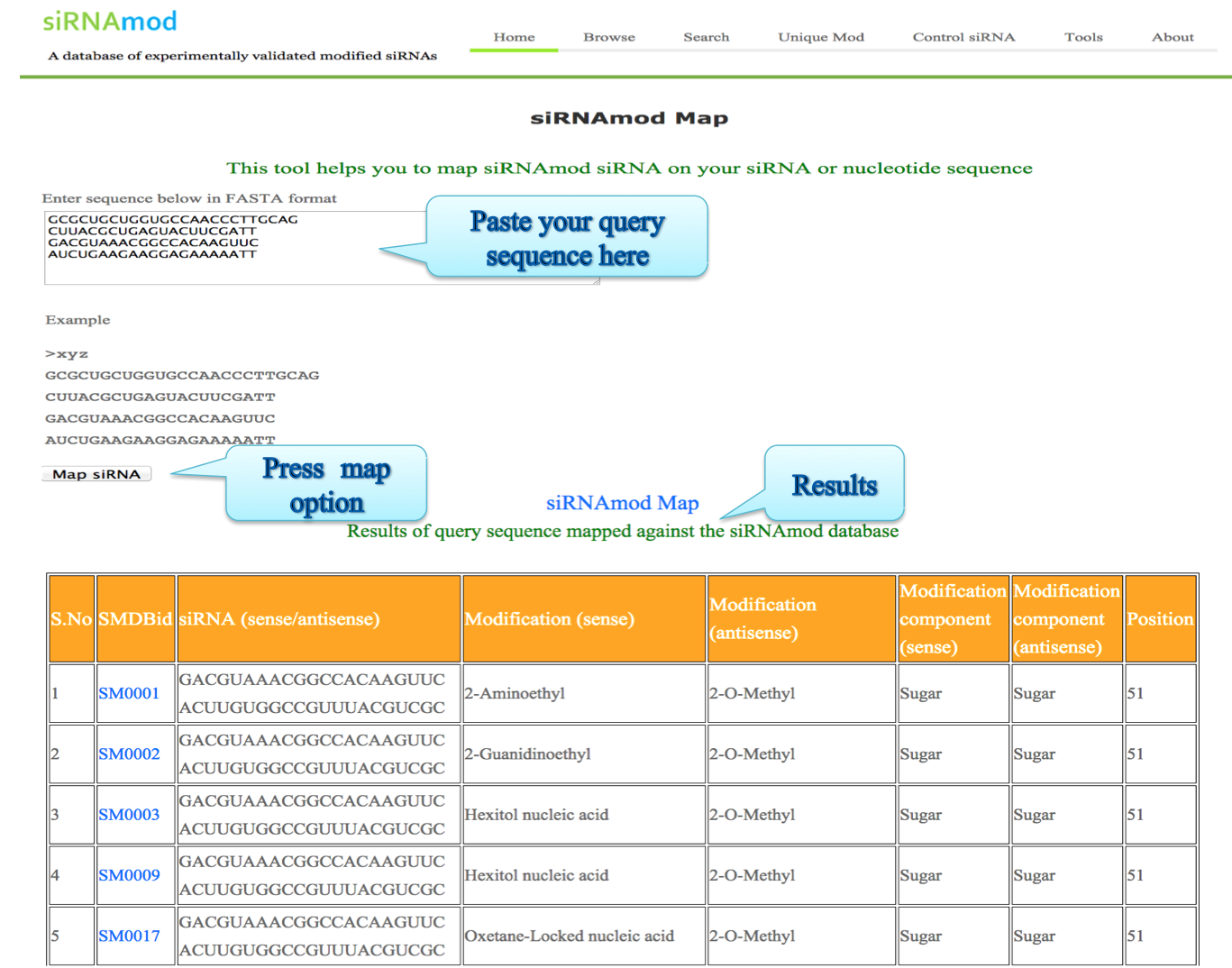
ii) Map:
This tool helps you to map database siRNAs on your siRNA or nucleotide sequence.
The output of this tool displays the SMBDid, modification in sense/antisense strand, modification position in sense/antisense strand, modified component in sense/antisense. Also mentioned is the start position where the match is found in the user provided sequence.