

This page provides help to the users of SATPdb on how to effectively use this dataset. For each module which is implemented in SATPdb, here we provide the figures and description on how to use that module. A user can click on the links given below to get help of the respective modules.
Browse | |
Function/SubFunction This module displays the distribution of peptides according to their function and subfunction. | |
 | |
| Result | |
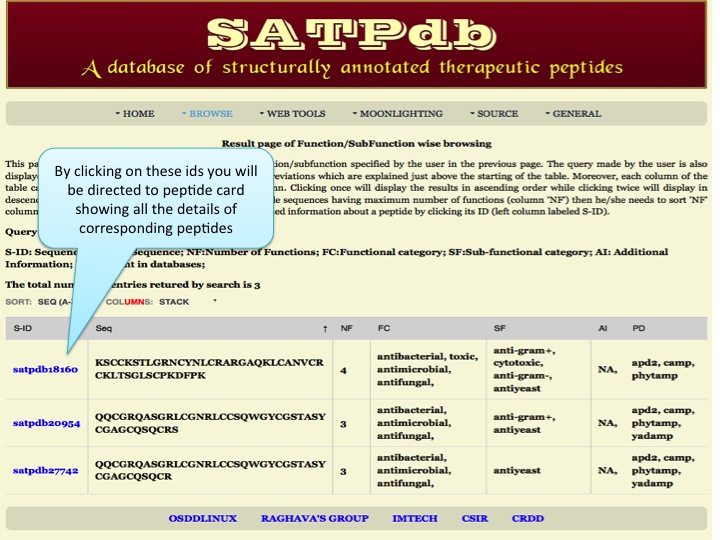 | |
Properties This page provides the distribution of peptides present in the SATPdb database based on the property of peptides such as Hydrophobic residues, Polar residues, Positively charged, Negatively charged and small sized residues. | |
 | |
| Result | |
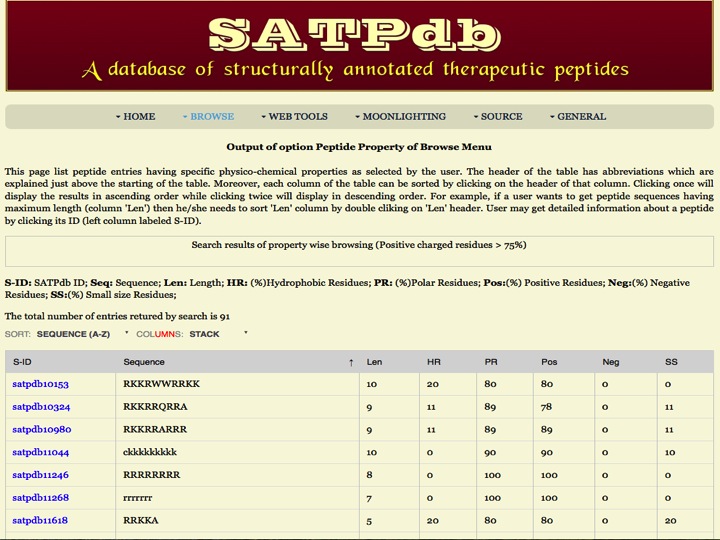 | |
Modification This page provides the distribution of different modifications (Terminal modifications, PTMs, non-natural residues) present in the peptides of SATPdb database. | |
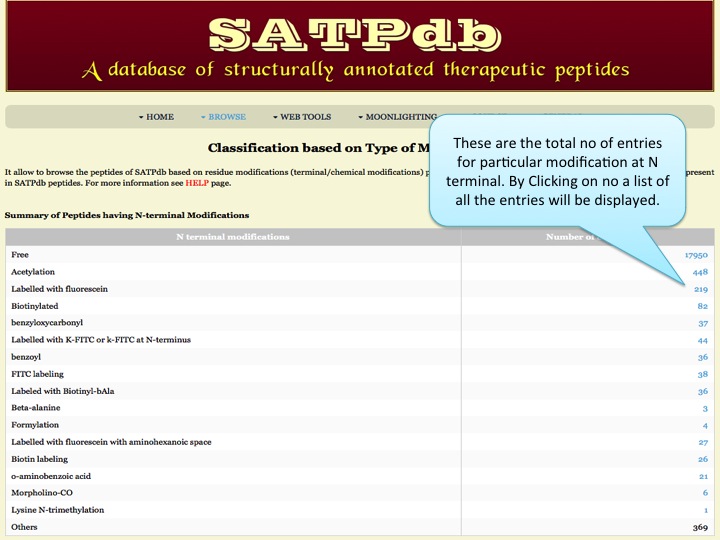 | |
| Result | |
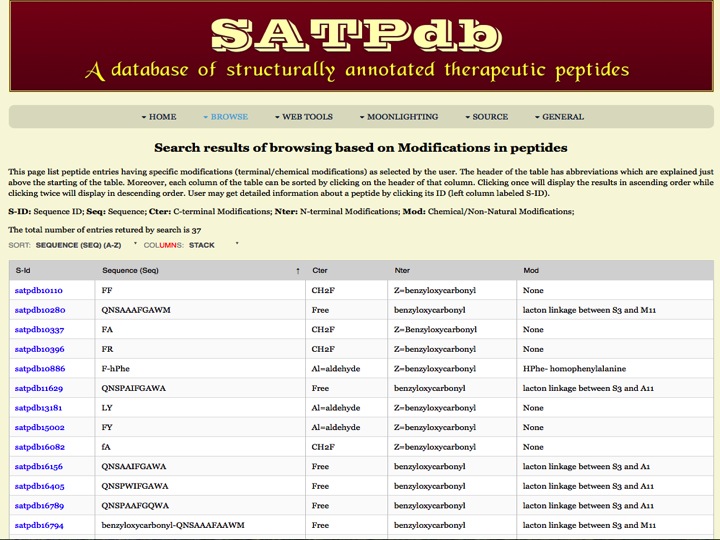 | |
Seconadary Structure Here the user can browse the peptides on the basis of their secondary structure elements like helix, strands, turns and coils. | |
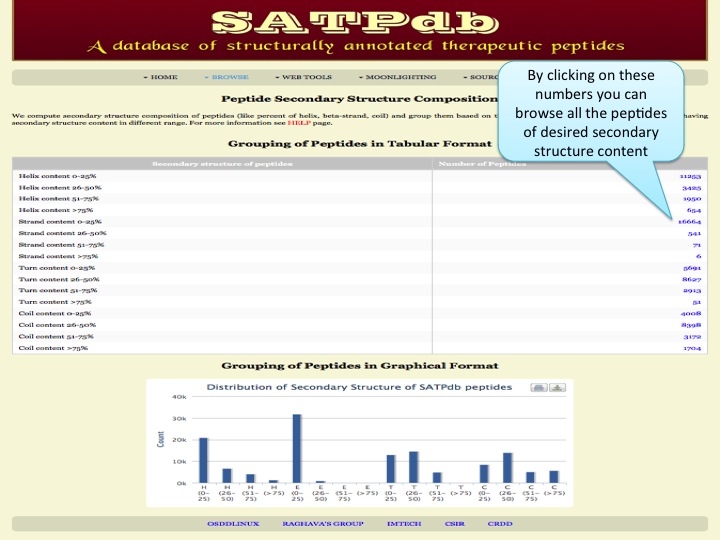 | Result |
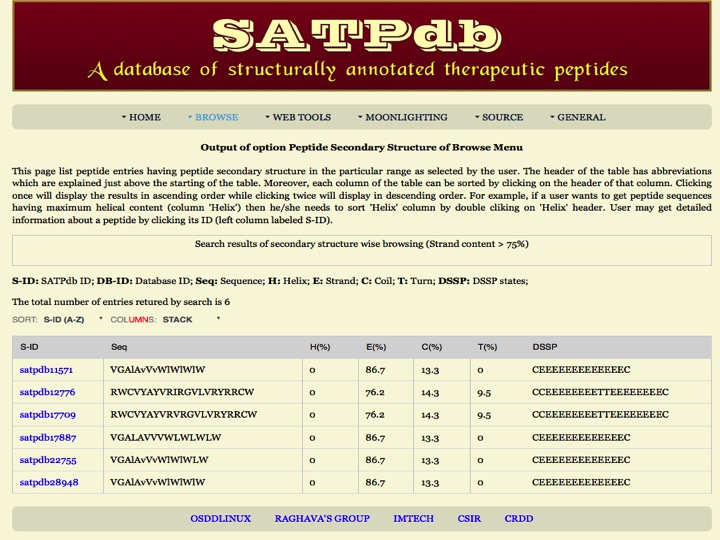 | |
Frequency Here the user can browse the peptides based on the frequency of their occurence in different databases. | |
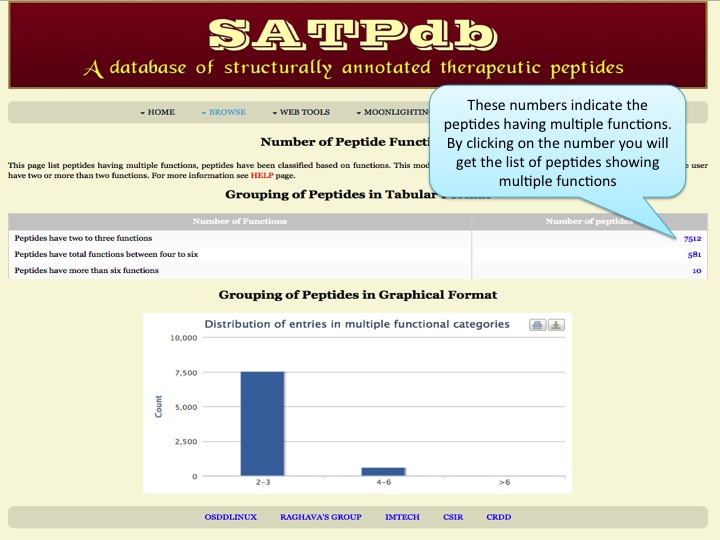 | |
| Result | |
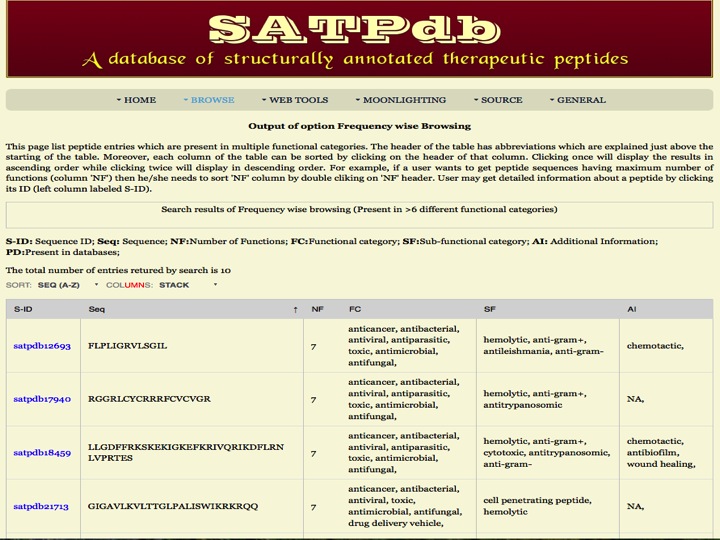 | |
Additional info Here the user can browse the peptides which are not classified in functional or any subfunctional category but have additional important properties. | |
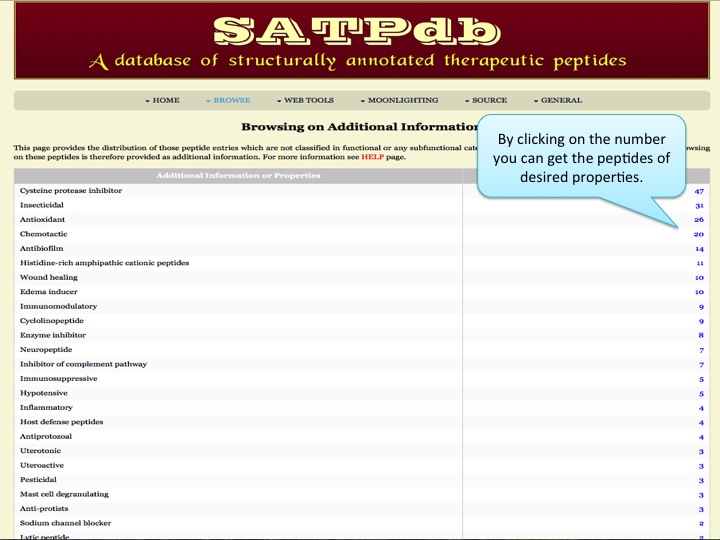 | |
Web Tools | |
Search This page allows users to search in any field or against multiple fields in SATPdb | |
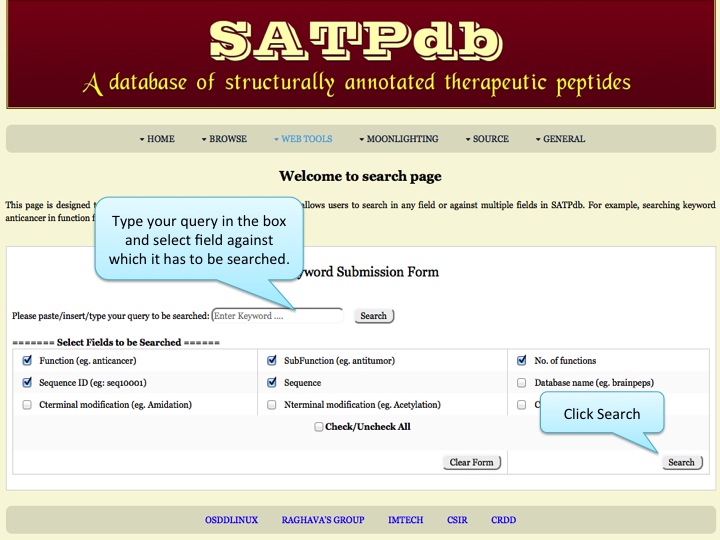 | |
| Result | |
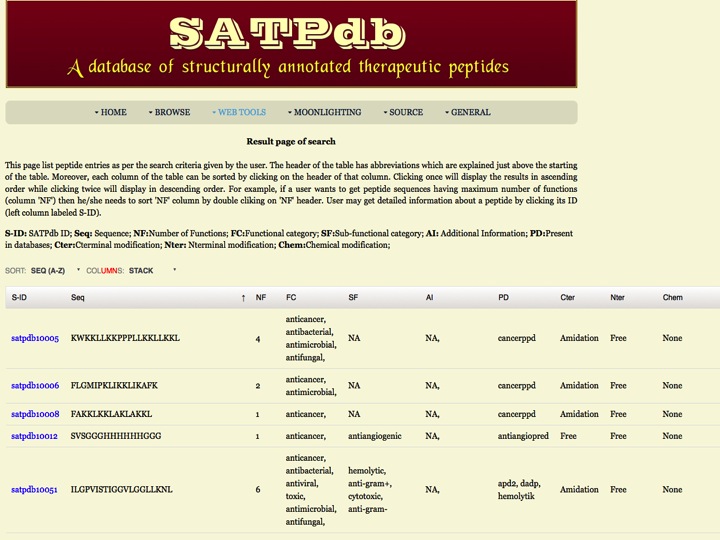 | |
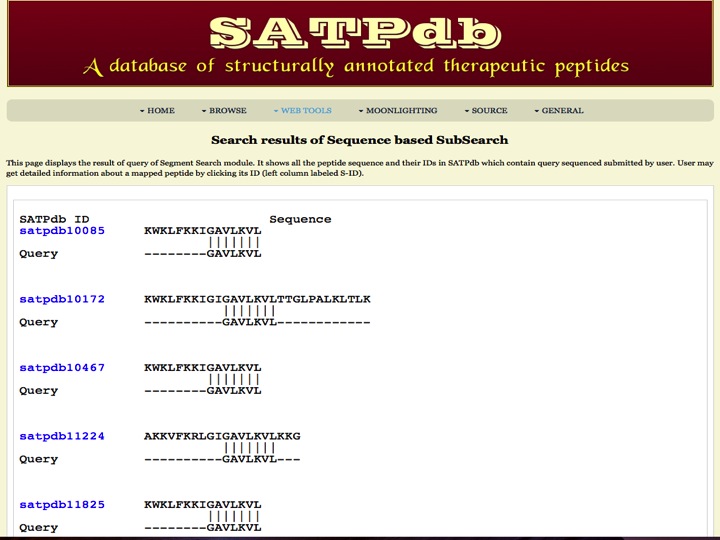
Segment Search This page allows users to search and map a peptide in all the databases covered in SATPdb. | |
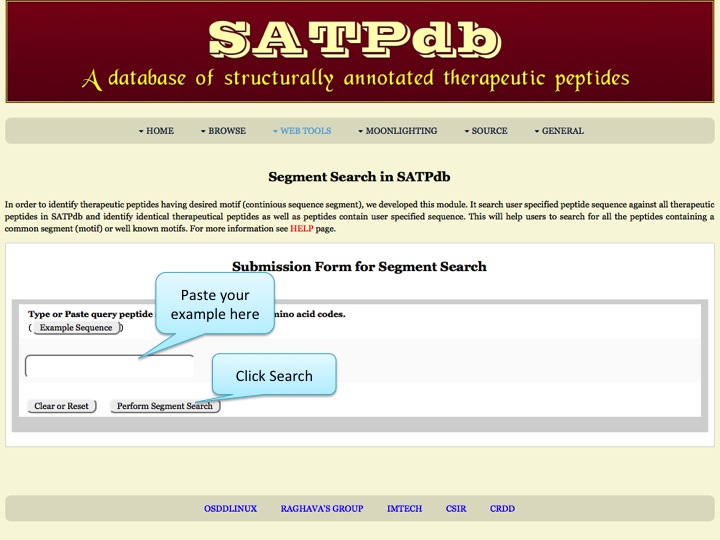 | |
| Result | |
 | |
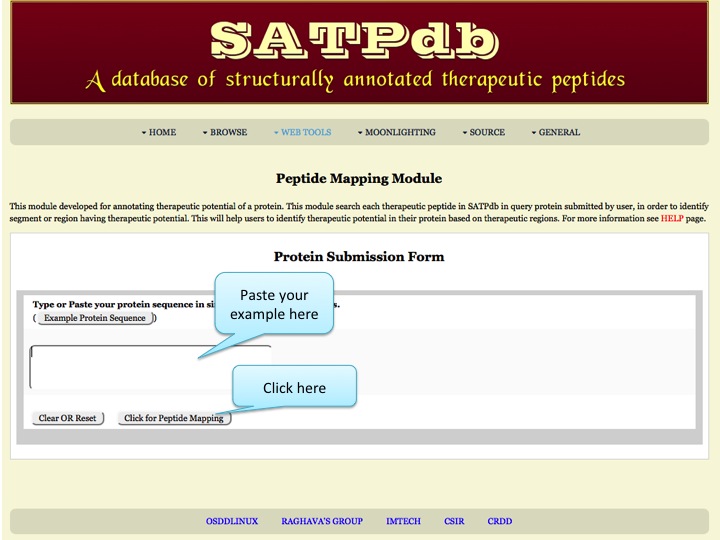
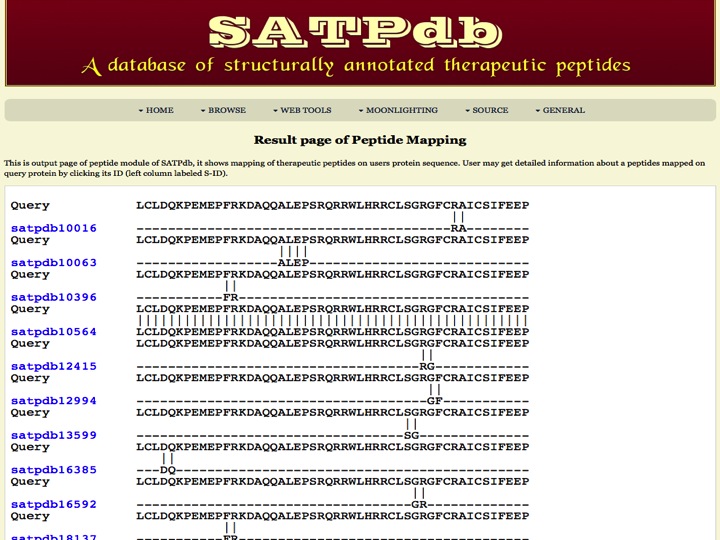
Peptide Mapping This page allows the users to map a protein in the repository of peptides from all the databases covered in SATPdb | |
 | |
| Result | |
 | |
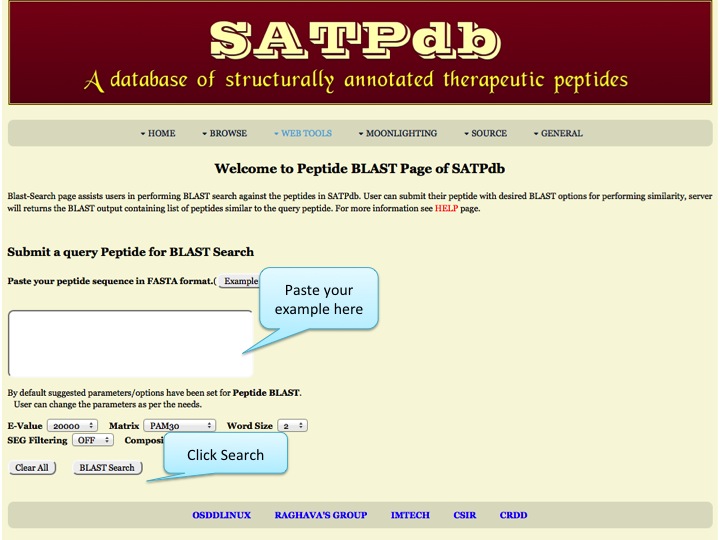
Sequence Similarity Blast-Search page assists users in performing BLAST search against the peptides in SATPdb. User can submit their peptide with desired BLAST options for performing similarity, server will returns the BLAST output containing list of peptides similar to the query peptide. | |
 | |
| Result | |
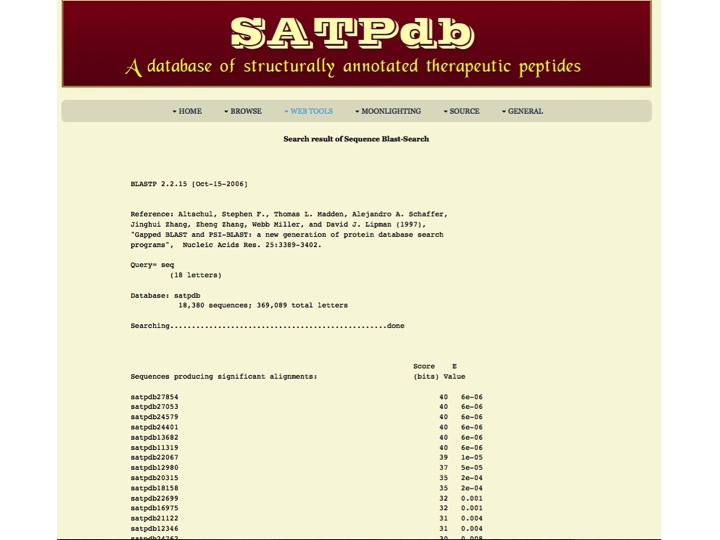 | |
Secondary Structure This page allows users to search and map the secondary structure of query peptide in all the entries covered in SATPdb. | |
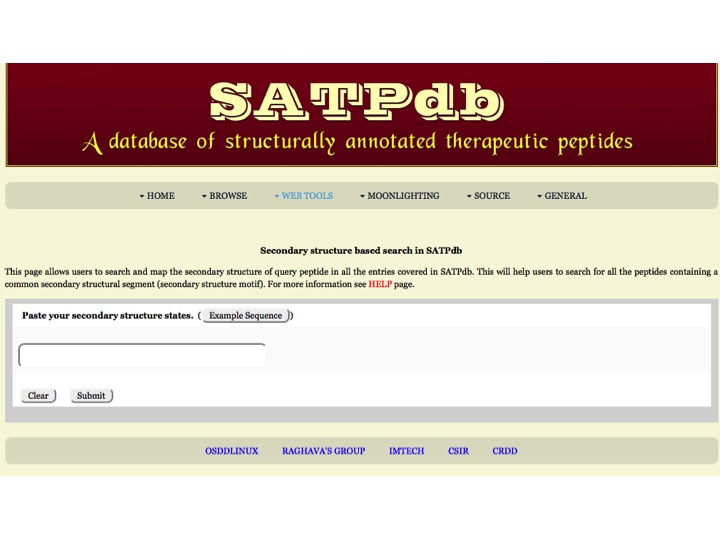 | |
Tertiary Structure This page has been created to search the peptides using structure input. The page can accept the input in major file formats (PDB, Smiles). User can also draw their stucture using Marvin applet. The page will further process the input file with the user-selected options using JCsearch program at the backend and the results will be provided in an attractive passion. The PDB input option is set to default. | |
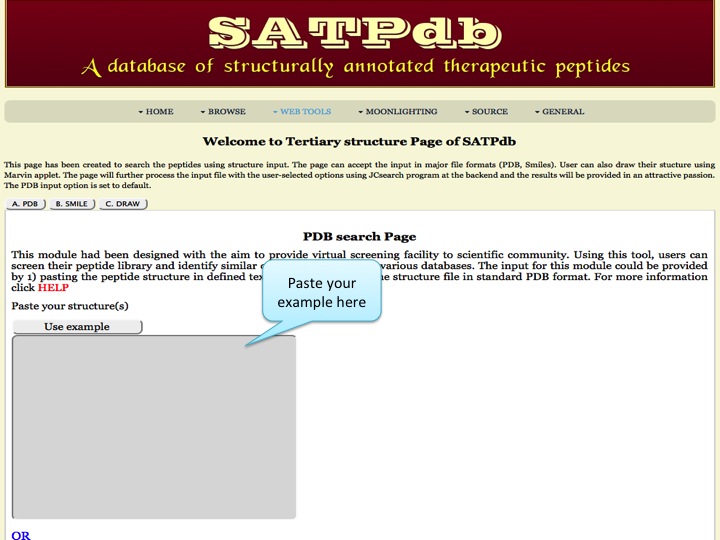 | |
| Result | |
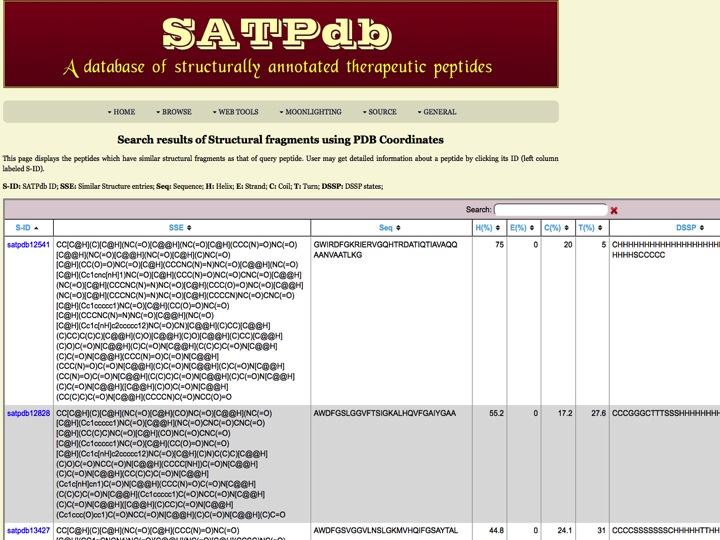 | |
Moonlighting | |
Desired Function This page allows users to search peptide(s) satisfying the conditional search based upon the desired properties defined by the user. User can use AND, OR and NOT boolean operators to select conditions based upon the question in his/her mind. Output will be displayed with the peptides satisfying that complex condition given by the user. | |
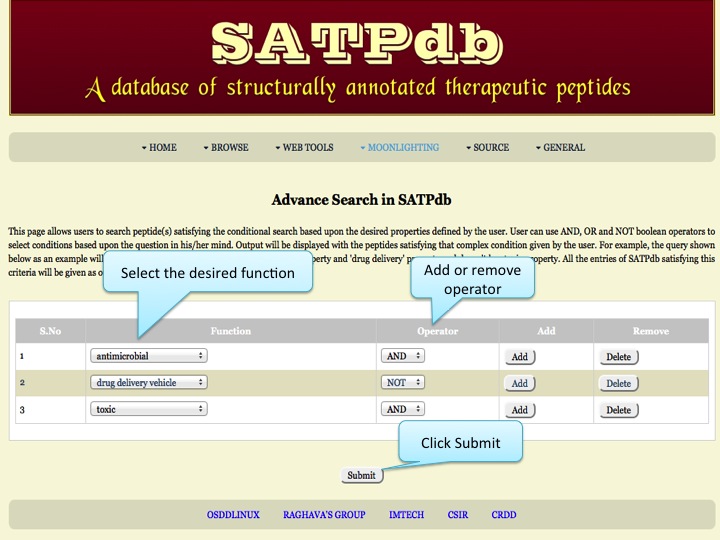 | |
| Result | |
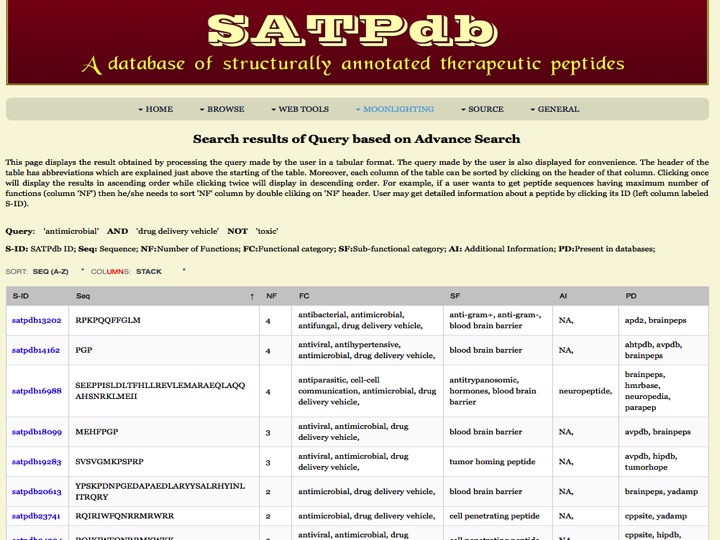 | |
Multiple Function This page allows the users to search peptides which are present in multiple functional categories selected by the user. | |
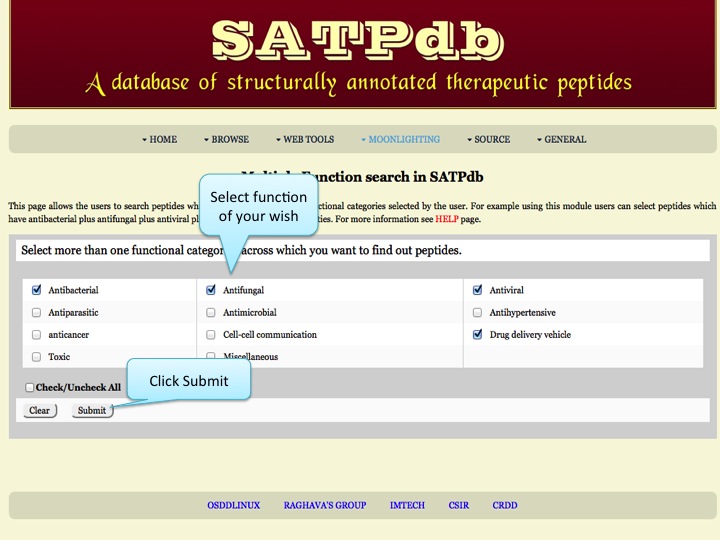 | |
| Result | |
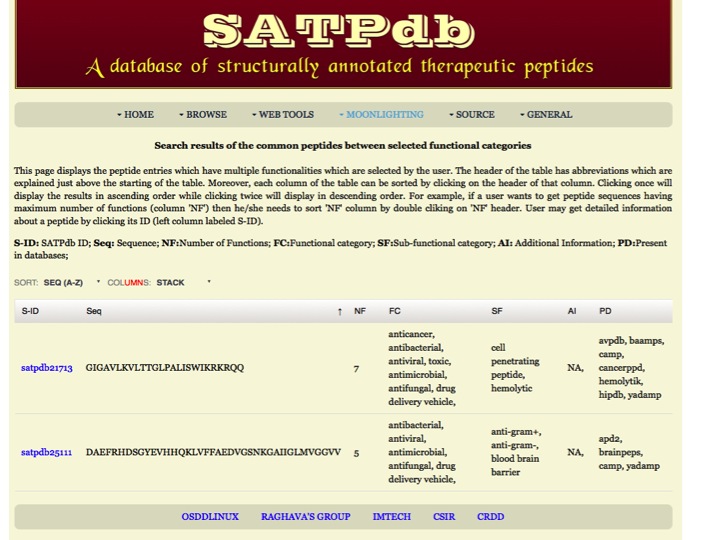 | |
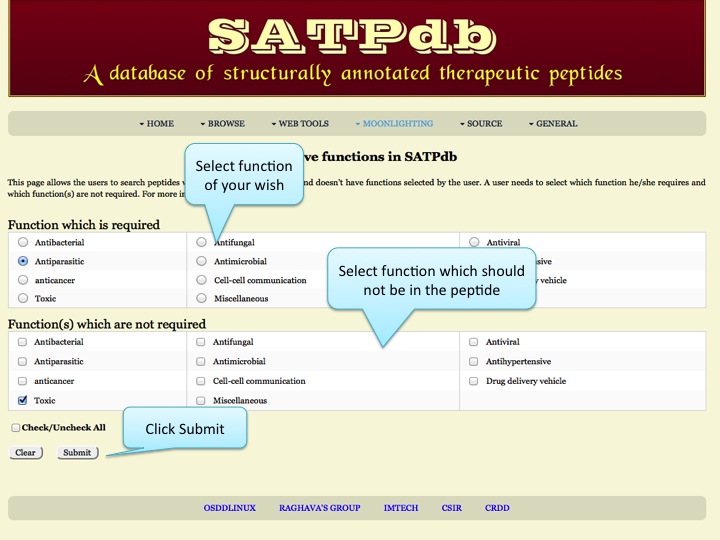
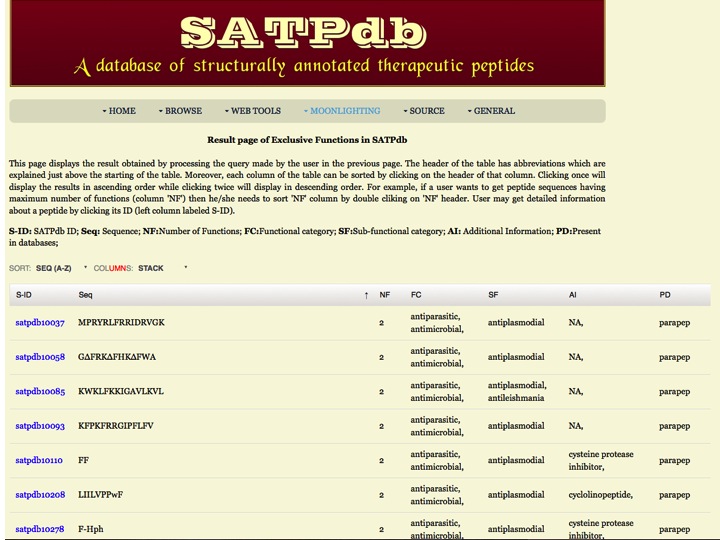
Exclusive Function This page allows the users to search peptides which have exclusive function and doesn't have functions selected by the user | |
 | |
| Result | |
 | |
| |
|